 | | PATHWAY: cak00903 | |
| Entry |
|
|---|
| Name |
Limonene degradation - Caulobacter sp. K31 |
|---|
| Class |
Metabolism; Metabolism of terpenoids and polyketides
|
|---|
| Pathway map |
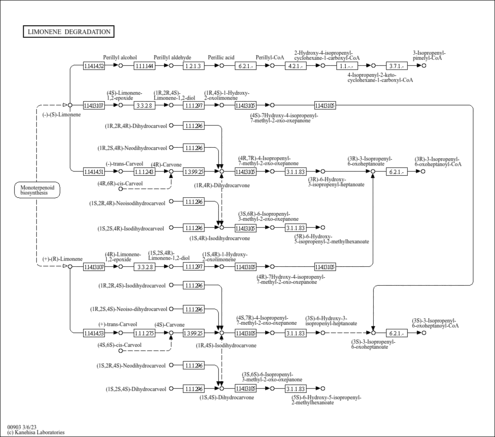
|
|---|
| Other DBs |
|
|---|
| Organism |
Caulobacter sp. K31 [GN: cak]
|
|---|
| Gene |
|
|---|
| Compound |
| C11396 | (1R,2R,4R)-Dihydrocarveol |
| C11397 | (1R,2S,4R)-Neo-dihydrocarveol |
| C11399 | (1S,2S,4R)-Iso-dihydrocarveol |
| C11400 | (1S,2R,4R)-Neoiso-dihydrocarveol |
| C11401 | (1S,4R)-Iso-dihydrocarvone |
| C11402 | (4R,7R)-4-Isopropenyl-7-methyl-2-oxo-oxepanone |
| C11403 | (3S,6R)-6-Isopropenyl-3-methyl-2-oxo-oxepanone |
| C11404 | (3R)-6-Hydroxy-3-isopropenyl-heptanoate |
| C11405 | (3R)-3-Isopropenyl-6-oxoheptanoate |
| C11406 | (5R)-6-Hydroxy-5-isopropenyl-2-methylhexanoate |
| C11407 | (3R)-3-Isopropenyl-6-oxoheptanoyl-CoA |
| C11410 | (1R,2S,4S)-Neoiso-dihydrocarveol |
| C11411 | (1R,2R,4S)-Iso-dihydrocarveol |
| C11412 | (1R,4S)-Iso-dihydrocarvone |
| C11413 | (1S,2S,4S)-Dihydrocarveol |
| C11414 | (4S,7R)-4-Isopropenyl-7-methyl-2-oxo-oxepanone |
| C11416 | (1S,2R,4S)-Neo-dihydrocarveol |
| C11417 | (3S)-6-Hydroxy-3-isopropenyl-heptanoate |
| C11418 | (3S,6S)-6-Isopropenyl-3-methyl-2-oxo-oxepanone |
| C11419 | (3S)-3-Isopropenyl-6-oxoheptanoate |
| C11420 | (5S)-6-Hydroxy-5-isopropenyl-2-methylhexanoate |
| C11421 | (3S)-3-Isopropenyl-6-oxoheptanoyl-CoA |
| C11934 | 2-Hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA |
| C11935 | 4-Isopropenyl-2-oxy-cyclohexanecarboxyl-CoA |
| C11936 | 3-Isopropenylpimelyl-CoA |
| C11937 | (1S,4R)-1-Hydroxy-2-oxolimonene |
| C19079 | (4R)-7-Hydroxy-4-isopropenyl-7-methyl-2-oxo-oxepanone |
| C19081 | (4S)-Limonene-1,2-epoxide |
| C19082 | (1R,2R,4S)-Limonene-1,2-diol |
| C19083 | (1R,4S)-1-Hydroxy-2-oxolimonene |
| C19084 | (4S)-7-Hydroxy-4-isopropenyl-7-methyl-2-oxo-oxepanone |
|
|---|
| Reference |
|
|---|
| Authors |
van der Werf MJ, Swarts HJ, de Bont JA |
|---|
| Title |
Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
van der Werf MJ, Boot AM |
|---|
| Title |
Metabolism of carveol and dihydrocarveol in Rhodococcus erythropolis DCL14. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
|
|
|