 | | PATHWAY: map01100 | |
| Entry |
|
|---|
| Name |
Metabolic pathways |
|---|
| Class |
|
|---|
| Pathway map |
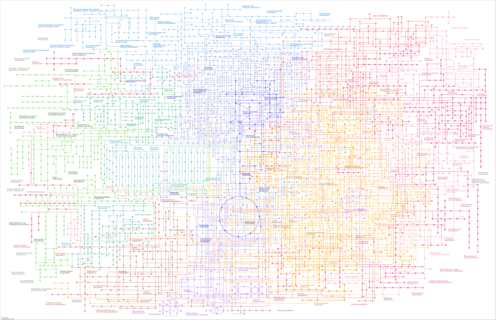
|
|---|
| Module |
| M00001 | Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate [PATH:map01100] |
| M00002 | Glycolysis, core module involving three-carbon compounds [PATH:map01100] |
| M00004 | Pentose phosphate pathway (Pentose phosphate cycle) [PATH:map01100] |
| M00006 | Pentose phosphate pathway, oxidative phase, glucose 6P => ribulose 5P [PATH:map01100] |
| M00007 | Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P [PATH:map01100] |
| M00008 | Entner-Doudoroff pathway, glucose-6P => glyceraldehyde-3P + pyruvate [PATH:map01100] |
| M00010 | Citrate cycle, first carbon oxidation, oxaloacetate => 2-oxoglutarate [PATH:map01100] |
| M00011 | Citrate cycle, second carbon oxidation, 2-oxoglutarate => oxaloacetate [PATH:map01100] |
| M00013 | Malonate semialdehyde pathway, propanoyl-CoA => acetyl-CoA [PATH:map01100] |
| M00016 | Lysine biosynthesis, succinyl-DAP pathway, aspartate => lysine [PATH:map01100] |
| M00017 | Methionine biosynthesis, aspartate => homoserine => methionine [PATH:map01100] |
| M00018 | Threonine biosynthesis, aspartate => homoserine => threonine [PATH:map01100] |
| M00019 | Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine [PATH:map01100] |
| M00022 | Shikimate pathway, phosphoenolpyruvate + erythrose-4P => chorismate [PATH:map01100] |
| M00023 | Tryptophan biosynthesis, chorismate => tryptophan [PATH:map01100] |
| M00024 | Phenylalanine biosynthesis, chorismate => phenylpyruvate => phenylalanine [PATH:map01100] |
| M00025 | Tyrosine biosynthesis, chorismate => HPP => tyrosine [PATH:map01100] |
| M00030 | Lysine biosynthesis, AAA pathway, 2-oxoglutarate => 2-aminoadipate => lysine [PATH:map01100] |
| M00031 | Lysine biosynthesis, mediated by LysW, 2-aminoadipate => lysine [PATH:map01100] |
| M00032 | Lysine degradation, lysine => saccharopine => acetoacetyl-CoA [PATH:map01100] |
| M00036 | Leucine degradation, leucine => acetoacetate + acetyl-CoA [PATH:map01100] |
| M00037 | Melatonin biosynthesis, animals, tryptophan => serotonin => melatonin [PATH:map01100] |
| M00038 | Tryptophan metabolism, tryptophan => kynurenine => 2-aminomuconate [PATH:map01100] |
| M00039 | Monolignol biosynthesis, phenylalanine/tyrosine => monolignol [PATH:map01100] |
| M00040 | Tyrosine biosynthesis, chorismate => arogenate => tyrosine [PATH:map01100] |
| M00042 | Catecholamine biosynthesis, tyrosine => dopamine => noradrenaline => adrenaline [PATH:map01100] |
| M00043 | Thyroid hormone biosynthesis, tyrosine => triiodothyronine/thyroxine [PATH:map01100] |
| M00044 | Tyrosine degradation, tyrosine => homogentisate [PATH:map01100] |
| M00045 | Histidine degradation, histidine => N-formiminoglutamate => glutamate [PATH:map01100] |
| M00046 | Pyrimidine degradation, uracil => beta-alanine, thymine => 3-aminoisobutanoate [PATH:map01100] |
| M00048 | De novo purine biosynthesis, PRPP + glutamine => IMP [PATH:map01100] |
| M00049 | Adenine ribonucleotide biosynthesis, IMP => ADP,ATP [PATH:map01100] |
| M00050 | Guanine ribonucleotide biosynthesis, IMP => GDP,GTP [PATH:map01100] |
| M00051 | De novo pyrimidine biosynthesis, glutamine (+ PRPP) => UMP [PATH:map01100] |
| M00052 | Pyrimidine ribonucleotide biosynthesis, UMP => UDP/UTP,CDP/CTP [PATH:map01100] |
| M00053 | Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP [PATH:map01100] |
| M00057 | Glycosaminoglycan biosynthesis, linkage tetrasaccharide [PATH:map01100] |
| M00058 | Glycosaminoglycan biosynthesis, chondroitin sulfate backbone [PATH:map01100] |
| M00059 | Glycosaminoglycan biosynthesis, heparan sulfate backbone [PATH:map01100] |
| M00060 | KDO2-lipid A biosynthesis, Raetz pathway, LpxL-LpxM type [PATH:map01100] |
| M00061 | D-Glucuronate degradation, D-glucuronate => pyruvate + D-glyceraldehyde 3P [PATH:map01100] |
| M00067 | Sulfoglycolipids biosynthesis, ceramide/1-alkyl-2-acylglycerol => sulfatide/seminolipid [PATH:map01100] |
| M00068 | Glycosphingolipid biosynthesis, globo-series, LacCer => Gb4Cer [PATH:map01100] |
| M00069 | Glycosphingolipid biosynthesis, ganglio series, LacCer => GT3 [PATH:map01100] |
| M00070 | Glycosphingolipid biosynthesis, lacto-series, LacCer => Lc4Cer [PATH:map01100] |
| M00071 | Glycosphingolipid biosynthesis, neolacto-series, LacCer => nLc4Cer [PATH:map01100] |
| M00088 | Ketone body biosynthesis, acetyl-CoA => acetoacetate/3-hydroxybutyrate/acetone [PATH:map01100] |
| M00090 | Phosphatidylcholine (PC) biosynthesis, choline => PC [PATH:map01100] |
| M00091 | Phosphatidylcholine (PC) biosynthesis, PE => PC [PATH:map01100] |
| M00092 | Phosphatidylethanolamine (PE) biosynthesis, ethanolamine => PE [PATH:map01100] |
| M00093 | Phosphatidylethanolamine (PE) biosynthesis, PA => PS => PE [PATH:map01100] |
| M00096 | C5 isoprenoid biosynthesis, non-mevalonate pathway [PATH:map01100] |
| M00097 | beta-Carotene biosynthesis, GGAP => beta-carotene [PATH:map01100] |
| M00102 | Ergocalciferol biosynthesis, FPP => ergosterol/ergocalciferol [PATH:map01100] |
| M00104 | Bile acid biosynthesis, cholesterol => cholate/chenodeoxycholate [PATH:map01100] |
| M00106 | Conjugated bile acid biosynthesis, cholate => taurocholate/glycocholate [PATH:map01100] |
| M00107 | Steroid hormone biosynthesis, cholesterol => pregnenolone => progesterone [PATH:map01100] |
| M00108 | C21-Steroid hormone biosynthesis, progesterone => corticosterone/aldosterone [PATH:map01100] |
| M00109 | C21-Steroid hormone biosynthesis, progesterone => cortisol/cortisone [PATH:map01100] |
| M00110 | C19/C18-Steroid hormone biosynthesis, pregnenolone => androstenedione => estrone [PATH:map01100] |
| M00112 | Tocopherol/tocotorienol biosynthesis, homogentisate + phytyl/geranylgeranyl-PP => tocopherol/tocotorienol [PATH:map01100] |
| M00114 | Ascorbate biosynthesis, plants, fructose-6P => ascorbate [PATH:map01100] |
| M00115 | NAD biosynthesis, aspartate => quinolinate => NAD [PATH:map01100] |
| M00116 | Menaquinone biosynthesis, chorismate (+ polyprenyl-PP) => menaquinol [PATH:map01100] |
| M00117 | Ubiquinone biosynthesis, prokaryotes, chorismate (+ polyprenyl-PP) => ubiquinol [PATH:map01100] |
| M00118 | Glutathione biosynthesis, glutamate => glutathione [PATH:map01100] |
| M00119 | Pantothenate biosynthesis, valine/L-aspartate => pantothenate [PATH:map01100] |
| M00121 | Heme biosynthesis, plants and bacteria, glutamate => heme [PATH:map01100] |
| M00122 | Cobalamin biosynthesis, cobyrinate a,c-diamide => cobalamin [PATH:map01100] |
| M00123 | Biotin biosynthesis, pimeloyl-ACP/CoA => biotin [PATH:map01100] |
| M00124 | Pyridoxal-P biosynthesis, erythrose-4P => pyridoxal-P [PATH:map01100] |
| M00125 | Riboflavin biosynthesis, plants and bacteria, GTP => riboflavin/FMN/FAD [PATH:map01100] |
| M00127 | Thiamine biosynthesis, prokaryotes, AIR (+ DXP/tyrosine) => TMP/TPP [PATH:map01100] |
| M00128 | Ubiquinone biosynthesis, eukaryotes, 4-hydroxybenzoate + polyprenyl-PP => ubiquinol [PATH:map01100] |
| M00129 | Ascorbate biosynthesis, animals, glucose-1P => ascorbate [PATH:map01100] |
| M00130 | Inositol phosphate metabolism, PI=> PIP2 => Ins(1,4,5)P3 => Ins(1,3,4,5)P4 [PATH:map01100] |
| M00131 | Inositol phosphate metabolism, Ins(1,3,4,5)P4 => Ins(1,3,4)P3 => myo-inositol [PATH:map01100] |
| M00132 | Inositol phosphate metabolism, Ins(1,3,4)P3 => phytate [PATH:map01100] |
| M00133 | Polyamine biosynthesis, arginine => agmatine => putrescine => spermidine [PATH:map01100] |
| M00134 | Polyamine biosynthesis, arginine => ornithine => putrescine [PATH:map01100] |
| M00135 | GABA biosynthesis, eukaryotes, putrescine => GABA [PATH:map01100] |
| M00136 | GABA biosynthesis, prokaryotes, putrescine => GABA [PATH:map01100] |
| M00137 | Flavanone biosynthesis, phenylalanine => naringenin [PATH:map01100] |
| M00138 | Flavonoid biosynthesis, naringenin => pelargonidin [PATH:map01100] |
| M00143 | NADH dehydrogenase (ubiquinone) Fe-S protein/flavoprotein complex, mitochondria [PATH:map01100] |
| M00145 | NAD(P)H:quinone oxidoreductase, chloroplasts and cyanobacteria [PATH:map01100] |
| M00146 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex [PATH:map01100] |
| M00147 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex [PATH:map01100] |
| M00165 | Reductive pentose phosphate cycle (Calvin cycle) [PATH:map01100] |
| M00170 | C4-dicarboxylic acid cycle, phosphoenolpyruvate carboxykinase type [PATH:map01100] |
| M00171 | C4-dicarboxylic acid cycle, NAD - malic enzyme type [PATH:map01100] |
| M00172 | C4-dicarboxylic acid cycle, NADP - malic enzyme type [PATH:map01100] |
| M00174 | Methane oxidation, methanotroph, methane => formaldehyde [PATH:map01100] |
| M00308 | Semi-phosphorylative Entner-Doudoroff pathway, gluconate => glycerate-3P [PATH:map01100] |
| M00309 | Non-phosphorylative Entner-Doudoroff pathway, gluconate/galactonate => glycerate [PATH:map01100] |
| M00338 | Cysteine biosynthesis, homocysteine + serine => cysteine [PATH:map01100] |
| M00344 | Formaldehyde assimilation, xylulose monophosphate pathway [PATH:map01100] |
| M00345 | Formaldehyde assimilation, ribulose monophosphate pathway [PATH:map01100] |
| M00367 | C10-C20 isoprenoid biosynthesis, non-plant eukaryotes [PATH:map01100] |
| M00369 | Cyanogenic glycoside biosynthesis, tyrosine => dhurrin [PATH:map01100] |
| M00370 | Glucosinolate biosynthesis, tryptophan => glucobrassicin [PATH:map01100] |
| M00371 | Castasterone biosynthesis, campesterol => castasterone [PATH:map01100] |
| M00372 | Abscisic acid biosynthesis, beta-carotene => abscisic acid [PATH:map01100] |
| M00377 | Reductive acetyl-CoA pathway (Wood-Ljungdahl pathway) [PATH:map01100] |
| M00418 | Toluene degradation, anaerobic, toluene => benzoyl-CoA [PATH:map01100] |
| M00432 | Leucine biosynthesis, 2-oxoisovalerate => 2-oxoisocaproate [PATH:map01100] |
| M00433 | Lysine biosynthesis, 2-oxoglutarate => 2-oxoadipate [PATH:map01100] |
| M00525 | Lysine biosynthesis, acetyl-DAP pathway, aspartate => lysine [PATH:map01100] |
| M00526 | Lysine biosynthesis, DAP dehydrogenase pathway, aspartate => lysine [PATH:map01100] |
| M00527 | Lysine biosynthesis, DAP aminotransferase pathway, aspartate => lysine [PATH:map01100] |
| M00530 | Dissimilatory nitrate reduction, nitrate => ammonia [PATH:map01100] |
| M00531 | Assimilatory nitrate reduction, nitrate => ammonia [PATH:map01100] |
| M00533 | Homoprotocatechuate degradation, homoprotocatechuate => 2-oxohept-3-enedioate [PATH:map01100] |
| M00534 | Naphthalene degradation, naphthalene => salicylate [PATH:map01100] |
| M00535 | Isoleucine biosynthesis, pyruvate => 2-oxobutanoate [PATH:map01100] |
| M00539 | Cumate degradation, p-cumate => 2-oxopent-4-enoate + 2-methylpropanoate [PATH:map01100] |
| M00540 | Benzoate degradation, cyclohexanecarboxylic acid =>pimeloyl-CoA [PATH:map01100] |
| M00541 | Benzoyl-CoA degradation, benzoyl-CoA => 3-hydroxypimeloyl-CoA [PATH:map01100] |
| M00543 | Biphenyl degradation, biphenyl => 2-oxopent-4-enoate + benzoate [PATH:map01100] |
| M00544 | Carbazole degradation, carbazole => 2-oxopent-4-enoate + anthranilate [PATH:map01100] |
| M00545 | Trans-cinnamate degradation, trans-cinnamate => acetyl-CoA [PATH:map01100] |
| M00547 | Benzene/toluene degradation, benzene => catechol / toluene => 3-methylcatechol [PATH:map01100] |
| M00549 | Nucleotide sugar biosynthesis, glucose => UDP-glucose [PATH:map01100] |
| M00550 | Ascorbate degradation, ascorbate => D-xylulose-5P [PATH:map01100] |
| M00551 | Benzoate degradation, benzoate => catechol / methylbenzoate => methylcatechol [PATH:map01100] |
| M00552 | D-galactonate degradation, De Ley-Doudoroff pathway, D-galactonate => glycerate-3P [PATH:map01100] |
| M00554 | Nucleotide sugar biosynthesis, galactose => UDP-galactose [PATH:map01100] |
| M00563 | Methanogenesis, methylamine/dimethylamine/trimethylamine => methane [PATH:map01100] |
| M00565 | Trehalose biosynthesis, D-glucose 1P => trehalose [PATH:map01100] |
| M00568 | Catechol ortho-cleavage, catechol => 3-oxoadipate [PATH:map01100] |
| M00569 | Catechol meta-cleavage, catechol => acetyl-CoA / 4-methylcatechol => propanoyl-CoA [PATH:map01100] |
| M00570 | Isoleucine biosynthesis, threonine => 2-oxobutanoate => isoleucine [PATH:map01100] |
| M00572 | Pimeloyl-ACP biosynthesis, BioC-BioH pathway, malonyl-ACP => pimeloyl-ACP [PATH:map01100] |
| M00573 | Biotin biosynthesis, BioI pathway, long-chain-acyl-ACP => pimeloyl-ACP => biotin [PATH:map01100] |
| M00577 | Biotin biosynthesis, BioW pathway, pimelate => pimeloyl-CoA => biotin [PATH:map01100] |
| M00579 | Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate [PATH:map01100] |
| M00580 | Pentose phosphate pathway, archaea, fructose 6P => ribose 5P [PATH:map01100] |
| M00595 | Thiosulfate oxidation by SOX complex, thiosulfate => sulfate [PATH:map01100] |
| M00596 | Dissimilatory sulfate reduction, sulfate => H2S [PATH:map01100] |
| M00608 | 2-Oxocarboxylic acid chain extension, 2-oxoglutarate => 2-oxoadipate => 2-oxopimelate => 2-oxosuberate [PATH:map01100] |
| M00620 | Incomplete reductive citrate cycle, acetyl-CoA => oxoglutarate [PATH:map01100] |
| M00623 | Phthalate degradation, phthalate => protocatechuate [PATH:map01100] |
| M00624 | Terephthalate degradation, terephthalate => 3,4-dihydroxybenzoate [PATH:map01100] |
| M00630 | D-Galacturonate degradation (fungi), D-galacturonate => glycerol [PATH:map01100] |
| M00631 | D-Galacturonate degradation (bacteria), D-galacturonate => pyruvate + D-glyceraldehyde 3P [PATH:map01100] |
| M00632 | Galactose degradation, Leloir pathway, galactose => alpha-D-glucose-1P [PATH:map01100] |
| M00633 | Semi-phosphorylative Entner-Doudoroff pathway, gluconate/galactonate => glycerate-3P [PATH:map01100] |
| M00636 | Phthalate degradation, phthalate => protocatechuate [PATH:map01100] |
| M00637 | Anthranilate degradation, anthranilate => catechol [PATH:map01100] |
| M00638 | Salicylate degradation, salicylate => gentisate [PATH:map01100] |
| M00661 | Paspaline biosynthesis, geranylgeranyl-PP + indoleglycerol phosphate => paspaline [PATH:map01100] |
| M00672 | Penicillin biosynthesis, aminoadipate + cycteine + valine => penicillin [PATH:map01100] |
| M00673 | Cephamycin C biosynthesis, aminoadipate + cycteine + valine => cephamycin C [PATH:map01100] |
| M00674 | Clavaminate biosynthesis, arginine + glyceraldehyde-3P => clavaminate [PATH:map01100] |
| M00675 | Carbapenem-3-carboxylate biosynthesis, pyrroline-5-carboxylate + malonyl-CoA => carbapenem-3-carboxylate [PATH:map01100] |
| M00736 | Nocardicin A biosynthesis, L-pHPG + arginine + serine => nocardicin A [PATH:map01100] |
| M00741 | Propanoyl-CoA metabolism, propanoyl-CoA => succinyl-CoA [PATH:map01100] |
| M00761 | Undecaprenylphosphate alpha-L-Ara4N biosynthesis, UDP-GlcA => undecaprenyl phosphate alpha-L-Ara4N [PATH:map01100] |
| M00763 | Ornithine biosynthesis, mediated by LysW, glutamate => ornithine [PATH:map01100] |
| M00773 | Tylosin biosynthesis, methylmalonyl-CoA + malonyl-CoA => tylactone => tylosin [PATH:map01100] |
| M00774 | Erythromycin biosynthesis, propanoyl-CoA + methylmalonyl-CoA => deoxyerythronolide B => erythromycin A/B [PATH:map01100] |
| M00775 | Oleandomycin biosynthesis, malonyl-CoA + methylmalonyl-CoA => 8,8a-deoxyoleandolide => oleandomycin [PATH:map01100] |
| M00776 | Pikromycin/methymycin biosynthesis, methylmalonyl-CoA + malonyl-CoA => narbonolide/10-deoxymethynolide => pikromycin/methymycin [PATH:map01100] |
| M00777 | Avermectin biosynthesis, 2-methylbutanoyl-CoA/isobutyryl-CoA => 6,8a-Seco-6,8a-deoxy-5-oxoavermectin 1a/1b aglycone => avermectin A1a/B1a/A1b/B1b [PATH:map01100] |
| M00778 | Type II polyketide backbone biosynthesis, acyl-CoA + malonyl-CoA => polyketide [PATH:map01100] |
| M00779 | Dihydrokalafungin biosynthesis, octaketide => dihydrokalafungin [PATH:map01100] |
| M00780 | Tetracycline/oxytetracycline biosynthesis, pretetramide => tetracycline/oxytetracycline [PATH:map01100] |
| M00781 | Nogalavinone/aklavinone biosynthesis, deoxynogalonate/deoxyaklanonate => nogalavinone/aklavinone [PATH:map01100] |
| M00782 | Mithramycin biosynthesis, 4-demethylpremithramycinone => mithramycin [PATH:map01100] |
| M00783 | Tetracenomycin C/8-demethyltetracenomycin C biosynthesis, tetracenomycin F2 => tetracenomycin C/8-demethyltetracenomycin C [PATH:map01100] |
| M00784 | Elloramycin biosynthesis, 8-demethyltetracenomycin C => elloramycin A [PATH:map01100] |
| M00785 | Cycloserine biosynthesis, arginine/serine => cycloserine [PATH:map01100] |
| M00786 | Fumitremorgin alkaloid biosynthesis, tryptophan + proline => fumitremorgin C/A [PATH:map01100] |
| M00787 | Bacilysin biosynthesis, prephenate => bacilysin [PATH:map01100] |
| M00789 | Rebeccamycin biosynthesis, tryptophan => rebeccamycin [PATH:map01100] |
| M00790 | Pyrrolnitrin biosynthesis, tryptophan => pyrrolnitrin [PATH:map01100] |
| M00804 | Complete nitrification, comammox, ammonia => nitrite => nitrate [PATH:map01100] |
| M00805 | Staurosporine biosynthesis, tryptophan => staurosporine [PATH:map01100] |
| M00808 | Violacein biosynthesis, tryptophan => violacein [PATH:map01100] |
| M00810 | Nicotine degradation, pyridine pathway, nicotine => 2,6-dihydroxypyridine/succinate semialdehyde [PATH:map01100] |
| M00811 | Nicotine degradation, pyrrolidine pathway, nicotine => succinate semialdehyde [PATH:map01100] |
| M00814 | Acarbose biosynthesis, sedoheptulopyranose-7P => acarbose [PATH:map01100] |
| M00815 | Validamycin A biosynthesis, sedoheptulopyranose-7P => validamycin A [PATH:map01100] |
| M00819 | Pentalenolactone biosynthesis, farnesyl-PP => pentalenolactone [PATH:map01100] |
| M00823 | Chlortetracycline biosynthesis, pretetramide => chlortetracycline [PATH:map01100] |
| M00824 | 9-membered enediyne core biosynthesis, malonyl-CoA => 3-hydroxyhexadeca-4,6,8,10,12,14-hexaenoyl-ACP => 9-membered enediyne core [PATH:map01100] |
| M00825 | 10-membered enediyne core biosynthesis, malonyl-CoA => 3-hydroxyhexadeca-4,6,8,10,12,14-hexaenoyl-ACP => 10-membered enediyne core [PATH:map01100] |
| M00826 | C-1027 benzoxazolinate moiety biosynthesis, chorismate => benzoxazolinyl-CoA [PATH:map01100] |
| M00827 | C-1027 beta-amino acid moiety biosynthesis, tyrosine => 3-chloro-4,5-dihydroxy-beta-phenylalanyl-PCP [PATH:map01100] |
| M00828 | Maduropeptin beta-hydroxy acid moiety biosynthesis, tyrosine => 3-(4-hydroxyphenyl)-3-oxopropanoyl-PCP [PATH:map01100] |
| M00829 | 3,6-Dimethylsalicylyl-CoA biosynthesis, malonyl-CoA => 6-methylsalicylate => 3,6-dimethylsalicylyl-CoA [PATH:map01100] |
| M00830 | Neocarzinostatin naphthoate moiety biosynthesis, malonyl-CoA => 2-hydroxy-5-methyl-1-naphthoate => 2-hydroxy-7-methoxy-5-methyl-1-naphthoyl-CoA [PATH:map01100] |
| M00831 | Kedarcidin 2-hydroxynaphthoate moiety biosynthesis, malonyl-CoA => 3,6,8-trihydroxy-2-naphthoate => 3-hydroxy-7,8-dimethoxy-6-isopropoxy-2-naphthoyl-CoA [PATH:map01100] |
| M00832 | Kedarcidin 2-aza-3-chloro-beta-tyrosine moiety biosynthesis, azatyrosine => 2-aza-3-chloro-beta-tyrosyl-PCP [PATH:map01100] |
| M00833 | Calicheamicin biosynthesis, calicheamicinone => calicheamicin [PATH:map01100] |
| M00834 | Calicheamicin orsellinate moiety biosynthesis, malonyl-CoA => orsellinate-ACP => 5-iodo-2,3-dimethoxyorsellinate-ACP [PATH:map01100] |
| M00835 | Pyocyanine biosynthesis, chorismate => pyocyanine [PATH:map01100] |
| M00836 | Coenzyme F430 biosynthesis, sirohydrochlorin => coenzyme F430 [PATH:map01100] |
| M00837 | Prodigiosin biosynthesis, L-proline => prodigiosin [PATH:map01100] |
| M00838 | Undecylprodigiosin biosynthesis, L-proline => undecylprodigiosin [PATH:map01100] |
| M00840 | Tetrahydrofolate biosynthesis, mediated by ribA and trpF, GTP => THF [PATH:map01100] |
| M00841 | Tetrahydrofolate biosynthesis, mediated by PTPS, GTP => THF [PATH:map01100] |
| M00843 | L-threo-Tetrahydrobiopterin biosynthesis, GTP => L-threo-BH4 [PATH:map01100] |
| M00845 | Arginine biosynthesis, glutamate => acetylcitrulline => arginine [PATH:map01100] |
| M00846 | Siroheme biosynthesis, glutamyl-tRNA => siroheme [PATH:map01100] |
| M00848 | Aurachin biosynthesis, anthranilate => aurachin A [PATH:map01100] |
| M00849 | C5 isoprenoid biosynthesis, mevalonate pathway, archaea [PATH:map01100] |
| M00854 | Glycogen biosynthesis, glucose-1P => glycogen/starch [PATH:map01100] |
| M00862 | beta-Oxidation, peroxisome, tri/dihydroxycholestanoyl-CoA => choloyl/chenodeoxycholoyl-CoA [PATH:map01100] |
| M00866 | KDO2-lipid A biosynthesis, Raetz pathway, non-LpxL-LpxM type [PATH:map01100] |
| M00868 | Heme biosynthesis, animals and fungi, glycine => heme [PATH:map01100] |
| M00873 | Fatty acid biosynthesis in mitochondria, animals [PATH:map01100] |
| M00875 | Staphyloferrin B biosynthesis, L-serine => staphyloferrin B [PATH:map01100] |
| M00876 | Staphyloferrin A biosynthesis, L-ornithine => staphyloferrin A [PATH:map01100] |
| M00877 | Kanosamine biosynthesis, glucose 6-phosphate => kanosamine [PATH:map01100] |
| M00878 | Phenylacetate degradation, phenylaxetate => acetyl-CoA/succinyl-CoA [PATH:map01100] |
| M00879 | Arginine succinyltransferase pathway, arginine => glutamate [PATH:map01100] |
| M00880 | Molybdenum cofactor biosynthesis, GTP => molybdenum cofactor [PATH:map01100] |
| M00881 | Lipoic acid biosynthesis, plants and bacteria, octanoyl-ACP => dihydrolipoyl-E2/H [PATH:map01100] |
| M00882 | Lipoic acid biosynthesis, eukaryotes, octanoyl-ACP => dihydrolipoyl-H [PATH:map01100] |
| M00883 | Lipoic acid biosynthesis, animals and bacteria, octanoyl-ACP => dihydrolipoyl-H => dihydrolipoyl-E2 [PATH:map01100] |
| M00884 | Lipoic acid biosynthesis, octanoyl-CoA => dihydrolipoyl-E2 [PATH:map01100] |
| M00891 | Ditryptophenaline biosynthesis, tryptophan + phenylalanine => ditryptophenaline [PATH:map01100] |
| M00892 | UDP-N-acetyl-D-glucosamine biosynthesis, eukaryotes, glucose => UDP-GlcNAc [PATH:map01100] |
| M00893 | Lovastatin biosynthesis, malonyl-CoA => lovastatin acid [PATH:map01100] |
| M00894 | Cannabidiol biosynthesis, malonyl-CoA => cannabidiol/dronabinol [PATH:map01100] |
| M00895 | Thiamine biosynthesis, prokaryotes, AIR (+ DXP/glycine) => TMP/TPP [PATH:map01100] |
| M00896 | Thiamine biosynthesis, archaea, AIR (+ NAD+) => TMP/TPP [PATH:map01100] |
| M00897 | Thiamine biosynthesis, plants, AIR (+ NAD+) => TMP/thiamine/TPP [PATH:map01100] |
| M00898 | Thiamine biosynthesis, pyridoxal-5P => TMP/thiamine/TPP [PATH:map01100] |
| M00901 | Fumiquinazoline biosynthesis, tryptophan + alanine + anthranilate => fumiquinazoline [PATH:map01100] |
| M00902 | Podophyllotoxin biosynthesis, coniferyl alcohol => podophyllotoxin [PATH:map01100] |
| M00903 | Fosfomycin biosynthesis, phosphoenolpyruvate => fosfomycin [PATH:map01100] |
| M00904 | Dapdiamides biosynthesis, L-2,3-diaminopropanoate => dapdiamide A/B/C [PATH:map01100] |
| M00905 | Grixazone biosynthesis, aspartate 4-semialdehyde => grixazone B [PATH:map01100] |
| M00906 | Ethynylserine biosynthesis, lysine => ethynylserine [PATH:map01100] |
| M00909 | UDP-N-acetyl-D-glucosamine biosynthesis, prokaryotes, glucose => UDP-GlcNAc [PATH:map01100] |
| M00910 | Phenylalanine biosynthesis, chorismate => arogenate => phenylalanine [PATH:map01100] |
| M00911 | Riboflavin biosynthesis, fungi, GTP => riboflavin/FMN/FAD [PATH:map01100] |
| M00912 | NAD biosynthesis, tryptophan => quinolinate => NAD [PATH:map01100] |
| M00913 | Pantothenate biosynthesis, 2-oxoisovalerate/spermine => pantothenate [PATH:map01100] |
| M00914 | Coenzyme A biosynthesis, archaea, 2-oxoisovalerate => 4-phosphopantoate => CoA [PATH:map01100] |
| M00916 | Pyridoxal-P biosynthesis, R5P + glyceraldehyde-3P + glutamine => pyridoxal-P [PATH:map01100] |
| M00917 | Phytosterol biosynthesis, squalene 2,3-epoxide => campesterol/sitosterol [PATH:map01100] |
| M00921 | Cyclooctatin biosynthesis, dimethylallyl-PP + isopentenyl-PP => cyclooctatin [PATH:map01100] |
| M00924 | Cobalamin biosynthesis, anaerobic, uroporphyrinogen III => sirohydrochlorin => cobyrinate a,c-diamide [PATH:map01100] |
| M00925 | Cobalamin biosynthesis, aerobic, uroporphyrinogen III => precorrin 2 => cobyrinate a,c-diamide [PATH:map01100] |
| M00926 | Heme biosynthesis, bacteria, glutamyl-tRNA => coproporphyrin III => heme [PATH:map01100] |
| M00928 | Gibberellin A4/A1 biosynthesis, GA12/GA53 => GA4/GA1 [PATH:map01100] |
| M00931 | Menaquinone biosynthesis, modified futalosine pathway [PATH:map01100] |
| M00932 | Phylloquinone biosynthesis, chorismate (+ phytyl-PP) => phylloquinol [PATH:map01100] |
| M00933 | Plastoquinone biosynthesis, homogentisate + solanesyl-PP => plastoquinol [PATH:map01100] |
| M00934 | Mycinamicin biosynthesis, malonyl-CoA + methylmalonyl-CoA => protomycinolide IV => mycinamicin II [PATH:map01100] |
| M00936 | Melatonin biosynthesis, plants, tryptophan => serotonin => melatonin [PATH:map01100] |
| M00937 | Aflatoxin biosynthesis, malonyl-CoA => aflatoxin B1 [PATH:map01100] |
| M00938 | Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP [PATH:map01100] |
| M00939 | Pyrimidine degradation, uracil => 3-hydroxypropanoate [PATH:map01100] |
| M00940 | Flavanone biosynthesis, p-coumaroyl-CoA => liquiritigenin [PATH:map01100] |
| M00941 | Isoflavone biosynthesis, liquiritigenin/naringenin => daidzein/genistein [PATH:map01100] |
| M00942 | Pterocarpan biosynthesis, daidzein => medicarpin [PATH:map01100] |
| M00943 | Reticuline biosynthesis, dopamine + 4HPAA => (S)-reticuline [PATH:map01100] |
| M00944 | Morphine biosynthesis, (S)-reticuline => morphine [PATH:map01100] |
| M00945 | Sanguinarine biosynthesis, (S)-reticuline => sanguinarine [PATH:map01100] |
| M00946 | Noscapine biosynthesis, (S)-reticuline => noscapine [PATH:map01100] |
| M00947 | D-Arginine racemization, D-arginine => L-arginine [PATH:map01100] |
| M00948 | Hydroxyproline degradation, trans-4-hydroxy-L-proline => 2-oxoglutarate [PATH:map01100] |
| M00949 | Staphylopine biosynthesis, L-histidine => staphylopine [PATH:map01100] |
| M00950 | Biotin biosynthesis, BioU pathway, pimeloyl-ACP/CoA => biotin [PATH:map01100] |
| M00951 | Cremeomycin biosynthesis, aspartate/3,4-AHBA => cremeomycin [PATH:map01100] |
| M00952 | Benzoxazinoid biosynthesis, indoleglycerol phosphate => DIMBOA-glucoside [PATH:map01100] |
| M00953 | Mugineic acid biosynthesis, methionine => 3-epihydroxymugineic acid [PATH:map01100] |
| M00956 | Lysine degradation, bacteria, L-lysine => succinate [PATH:map01100] |
| M00957 | Lysine degradation, bacteria, L-lysine => glutarate => succinate/acetyl-CoA [PATH:map01100] |
| M00958 | Adenine ribonucleotide degradation, AMP => Urate [PATH:map01100] |
| M00959 | Guanine ribonucleotide degradation, GMP => Urate [PATH:map01100] |
| M00960 | Lysine degradation, bacteria, L-lysine => D-lysine => succinate [PATH:map01100] |
| M00961 | Betacyanin biosynthesis, L-tyrosine => amaranthin [PATH:map01100] |
| M00962 | Psilocybin biosynthesis, tryptophan => psilocybin [PATH:map01100] |
| M00963 | Chanoclavine aldehyde biosynthesis, tryptophan => chanoclavine-I aldehyde [PATH:map01100] |
| M00964 | Fumigaclavine biosynthesis, chanoclavine-I aldehyde => fumigaclavine C [PATH:map01100] |
| M00965 | Vindoline biosynthesis, tabersonine => vindoline [PATH:map01100] |
| M00967 | Flavone degradation, luteolin/apigenin => DHCA/phloretate [PATH:map01100] |
| M00968 | Pentose bisphosphate pathway (nucleoside degradation), archaea, nucleoside/NMP => 3-PGA/glycerone phosphate [PATH:map01100] |
| M00969 | Fumagillin biosynthesis, farnesyl-PP => fumagillin [PATH:map01100] |
| M00974 | Betaine metabolism, animals, betaine => glycine [PATH:map01100] |
| M00975 | Betaine degradation, bacteria, betaine => pyruvate [PATH:map01100] |
| M00976 | C19-Steroid hormone biosynthesis, pregnenolone => testosterone => dihydrotestosterone [PATH:map01100] |
| M00977 | C19-Steroid hormone biosynthesis (androgen backdoor pathway), pregnenolone => androsterone => dihydrotestosterone [PATH:map01100] |
|
|---|
Related
pathway |
| map00040 | Pentose and glucuronate interconversions |
| map00051 | Fructose and mannose metabolism |
| map00053 | Ascorbate and aldarate metabolism |
| map00073 | Cutin, suberine and wax biosynthesis |
| map00121 | Secondary bile acid biosynthesis |
| map00130 | Ubiquinone and other terpenoid-quinone biosynthesis |
| map00196 | Photosynthesis - antenna proteins |
| map00250 | Alanine, aspartate and glutamate metabolism |
| map00260 | Glycine, serine and threonine metabolism |
| map00270 | Cysteine and methionine metabolism |
| map00280 | Valine, leucine and isoleucine degradation |
| map00290 | Valine, leucine and isoleucine biosynthesis |
| map00311 | Penicillin and cephalosporin biosynthesis |
| map00330 | Arginine and proline metabolism |
| map00361 | Chlorocyclohexane and chlorobenzene degradation |
| map00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| map00403 | Indole diterpene alkaloid biosynthesis |
| map00430 | Taurine and hypotaurine metabolism |
| map00440 | Phosphonate and phosphinate metabolism |
| map00512 | Mucin type O-glycan biosynthesis |
| map00513 | Various types of N-glycan biosynthesis |
| map00514 | Other types of O-glycan biosynthesis |
| map00515 | Mannose type O-glycan biosynthesis |
| map00520 | Amino sugar and nucleotide sugar metabolism |
| map00522 | Biosynthesis of 12-, 14- and 16-membered macrolides |
| map00523 | Polyketide sugar unit biosynthesis |
| map00524 | Neomycin, kanamycin and gentamicin biosynthesis |
| map00525 | Acarbose and validamycin biosynthesis |
| map00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate |
| map00533 | Glycosaminoglycan biosynthesis - keratan sulfate |
| map00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin |
| map00540 | Lipopolysaccharide biosynthesis |
| map00541 | O-Antigen nucleotide sugar biosynthesis |
| map00542 | O-Antigen repeat unit biosynthesis |
| map00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis |
| map00571 | Lipoarabinomannan (LAM) biosynthesis |
| map00572 | Arabinogalactan biosynthesis - Mycobacterium |
| map00592 | alpha-Linolenic acid metabolism |
| map00601 | Glycosphingolipid biosynthesis - lacto and neolacto series |
| map00603 | Glycosphingolipid biosynthesis - globo and isoglobo series |
| map00604 | Glycosphingolipid biosynthesis - ganglio series |
| map00624 | Polycyclic aromatic hydrocarbon degradation |
| map00625 | Chloroalkane and chloroalkene degradation |
| map00630 | Glyoxylate and dicarboxylate metabolism |
| map00660 | C5-Branched dibasic acid metabolism |
| map00710 | Carbon fixation in photosynthetic organisms |
| map00720 | Carbon fixation pathways in prokaryotes |
| map00760 | Nicotinate and nicotinamide metabolism |
| map00770 | Pantothenate and CoA biosynthesis |
| map00900 | Terpenoid backbone biosynthesis |
| map00907 | Pinene, camphor and geraniol degradation |
| map00909 | Sesquiterpenoid and triterpenoid biosynthesis |
| map00944 | Flavone and flavonol biosynthesis |
| map00950 | Isoquinoline alkaloid biosynthesis |
| map00960 | Tropane, piperidine and pyridine alkaloid biosynthesis |
| map00980 | Metabolism of xenobiotics by cytochrome P450 |
| map00982 | Drug metabolism - cytochrome P450 |
| map00983 | Drug metabolism - other enzymes |
| map00996 | Biosynthesis of various alkaloids |
| map00997 | Biosynthesis of various other secondary metabolites |
| map00998 | Biosynthesis of various antibiotics |
| map00999 | Biosynthesis of various plant secondary metabolites |
| map01040 | Biosynthesis of unsaturated fatty acids |
| map01053 | Biosynthesis of siderophore group nonribosomal peptides |
| map01055 | Biosynthesis of vancomycin group antibiotics |
| map01056 | Biosynthesis of type II polyketide backbone |
| map01057 | Biosynthesis of type II polyketide products |
| map01059 | Biosynthesis of enediyne antibiotics |
|
|---|
| KO pathway |
|
|---|
|
|
|