 | | PATHWAY: map01120 | |
| Entry |
|
|---|
| Name |
Microbial metabolism in diverse environments |
|---|
| Class |
|
|---|
| Pathway map |
| map01120 | Microbial metabolism in diverse environments |
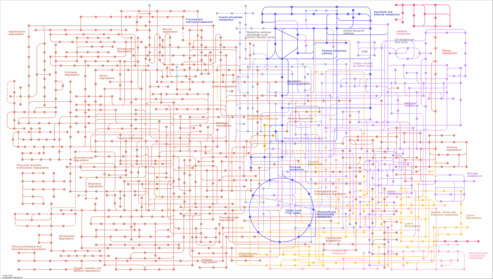
|
|---|
| Module |
| M00004 | Pentose phosphate pathway (Pentose phosphate cycle) [PATH:map01120] |
| M00006 | Pentose phosphate pathway, oxidative phase, glucose 6P => ribulose 5P [PATH:map01120] |
| M00007 | Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P [PATH:map01120] |
| M00008 | Entner-Doudoroff pathway, glucose-6P => glyceraldehyde-3P + pyruvate [PATH:map01120] |
| M00165 | Reductive pentose phosphate cycle (Calvin cycle) [PATH:map01120] |
| M00170 | C4-dicarboxylic acid cycle, phosphoenolpyruvate carboxykinase type [PATH:map01120] |
| M00171 | C4-dicarboxylic acid cycle, NAD - malic enzyme type [PATH:map01120] |
| M00172 | C4-dicarboxylic acid cycle, NADP - malic enzyme type [PATH:map01120] |
| M00174 | Methane oxidation, methanotroph, methane => formaldehyde [PATH:map01120] |
| M00308 | Semi-phosphorylative Entner-Doudoroff pathway, gluconate => glycerate-3P [PATH:map01120] |
| M00309 | Non-phosphorylative Entner-Doudoroff pathway, gluconate/galactonate => glycerate [PATH:map01120] |
| M00344 | Formaldehyde assimilation, xylulose monophosphate pathway [PATH:map01120] |
| M00345 | Formaldehyde assimilation, ribulose monophosphate pathway [PATH:map01120] |
| M00377 | Reductive acetyl-CoA pathway (Wood-Ljungdahl pathway) [PATH:map01120] |
| M00418 | Toluene degradation, anaerobic, toluene => benzoyl-CoA [PATH:map01120] |
| M00530 | Dissimilatory nitrate reduction, nitrate => ammonia [PATH:map01120] |
| M00531 | Assimilatory nitrate reduction, nitrate => ammonia [PATH:map01120] |
| M00533 | Homoprotocatechuate degradation, homoprotocatechuate => 2-oxohept-3-enedioate [PATH:map01120] |
| M00534 | Naphthalene degradation, naphthalene => salicylate [PATH:map01120] |
| M00539 | Cumate degradation, p-cumate => 2-oxopent-4-enoate + 2-methylpropanoate [PATH:map01120] |
| M00540 | Benzoate degradation, cyclohexanecarboxylic acid =>pimeloyl-CoA [PATH:map01120] |
| M00541 | Benzoyl-CoA degradation, benzoyl-CoA => 3-hydroxypimeloyl-CoA [PATH:map01120] |
| M00543 | Biphenyl degradation, biphenyl => 2-oxopent-4-enoate + benzoate [PATH:map01120] |
| M00544 | Carbazole degradation, carbazole => 2-oxopent-4-enoate + anthranilate [PATH:map01120] |
| M00545 | Trans-cinnamate degradation, trans-cinnamate => acetyl-CoA [PATH:map01120] |
| M00547 | Benzene/toluene degradation, benzene => catechol / toluene => 3-methylcatechol [PATH:map01120] |
| M00550 | Ascorbate degradation, ascorbate => D-xylulose-5P [PATH:map01120] |
| M00551 | Benzoate degradation, benzoate => catechol / methylbenzoate => methylcatechol [PATH:map01120] |
| M00563 | Methanogenesis, methylamine/dimethylamine/trimethylamine => methane [PATH:map01120] |
| M00568 | Catechol ortho-cleavage, catechol => 3-oxoadipate [PATH:map01120] |
| M00569 | Catechol meta-cleavage, catechol => acetyl-CoA / 4-methylcatechol => propanoyl-CoA [PATH:map01120] |
| M00579 | Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate [PATH:map01120] |
| M00580 | Pentose phosphate pathway, archaea, fructose 6P => ribose 5P [PATH:map01120] |
| M00595 | Thiosulfate oxidation by SOX complex, thiosulfate => sulfate [PATH:map01120] |
| M00596 | Dissimilatory sulfate reduction, sulfate => H2S [PATH:map01120] |
| M00608 | 2-Oxocarboxylic acid chain extension, 2-oxoglutarate => 2-oxoadipate => 2-oxopimelate => 2-oxosuberate [PATH:map01120] |
| M00620 | Incomplete reductive citrate cycle, acetyl-CoA => oxoglutarate [PATH:map01120] |
| M00623 | Phthalate degradation, phthalate => protocatechuate [PATH:map01120] |
| M00624 | Terephthalate degradation, terephthalate => 3,4-dihydroxybenzoate [PATH:map01120] |
| M00636 | Phthalate degradation, phthalate => protocatechuate [PATH:map01120] |
| M00637 | Anthranilate degradation, anthranilate => catechol [PATH:map01120] |
| M00638 | Salicylate degradation, salicylate => gentisate [PATH:map01120] |
| M00804 | Complete nitrification, comammox, ammonia => nitrite => nitrate [PATH:map01120] |
| M00810 | Nicotine degradation, pyridine pathway, nicotine => 2,6-dihydroxypyridine/succinate semialdehyde [PATH:map01120] |
| M00811 | Nicotine degradation, pyrrolidine pathway, nicotine => succinate semialdehyde [PATH:map01120] |
| M00836 | Coenzyme F430 biosynthesis, sirohydrochlorin => coenzyme F430 [PATH:map01120] |
| M00878 | Phenylacetate degradation, phenylaxetate => acetyl-CoA/succinyl-CoA [PATH:map01120] |
| M00956 | Lysine degradation, bacteria, L-lysine => succinate [PATH:map01120] |
| M00957 | Lysine degradation, bacteria, L-lysine => glutarate => succinate/acetyl-CoA [PATH:map01120] |
| M00960 | Lysine degradation, bacteria, L-lysine => D-lysine => succinate [PATH:map01120] |
|
|---|
Related
pathway |
| map00051 | Fructose and mannose metabolism |
| map00053 | Ascorbate and aldarate metabolism |
| map00260 | Glycine, serine and threonine metabolism |
| map00361 | Chlorocyclohexane and chlorobenzene degradation |
| map00440 | Phosphonate and phosphinate metabolism |
| map00624 | Polycyclic aromatic hydrocarbon degradation |
| map00625 | Chloroalkane and chloroalkene degradation |
| map00630 | Glyoxylate and dicarboxylate metabolism |
| map00710 | Carbon fixation by Calvin cycle |
| map00760 | Nicotinate and nicotinamide metabolism |
| map00907 | Pinene, camphor and geraniol degradation |
|
|---|
| KO pathway |
|
|---|
|
|
|