| Entry |
|
|---|
| Name |
Polyketide sugar unit biosynthesis |
|---|
| Class |
Metabolism; Metabolism of terpenoids and polyketides
|
|---|
| Pathway map |
| rn00523 | Polyketide sugar unit biosynthesis |
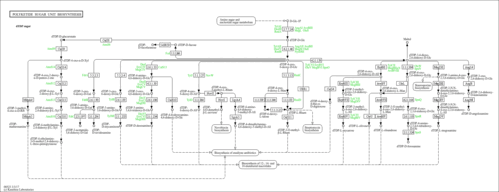
|
|---|
| Module |
| M00793 | dTDP-L-rhamnose biosynthesis, glucose-1P => dTDP-L-Rha [PATH:rn00523] |
|
|---|
| Reaction |
| R02328 | dTTP:alpha-D-glucose-1-phosphate thymidylyltransferase |
| R02773 | dTDP-4-amino-4,6-dideoxy-D-glucose:2-oxoglutarate aminotransferase |
| R02776 | dTDP-6-deoxy-L-talose:NADP+ 4-oxidoreductase |
| R02777 | dTDP-beta-L-rhamnose:NADP+ 4-oxidoreductase; dTDP-6-deoxy-L-mannose:NADP+ 4-oxidoreductase |
| R05518 | dTDP-4-dehydro-6-deoxy-alpha-D-glucopyranose hydro-lyase (dTDP-2,6-dideoxy-D-glycero-hex-2-enos-4-ulose-forming) |
| R05526 | dTDP-4-oxo-2,6-dideoxy-D-glucose:NADP+ 3-oxidoreductase |
| R05687 | dTDP-4-keto-6-deoxyglucose reductase |
| R06423 | dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose:2-oxoglutarate aminotransferase |
| R06424 | dTDP-4-dehydro-6-deoxy-alpha-D-glucopyranose:dTDP-3-dehydro-6-deoxy-alpha-D-glucopyranose isomerase |
| R06426 | dTDP-3-amino-3,4,6-trideoxy-D-glucose:2-oxoglutarate aminotransferase |
| R06427 | S-adenosyl-L-methionine:dTDP-3-amino-3,4,6-trideoxy-alpha-D-glucopyranose 3-N,N-dimethyltransferase |
| R06433 | dTDP-4-oxo-2,6-dideoxy-D-allose 5-epimerase |
| R06436 | dTDP-L-mycarose:NADP+ 4-oxidoreductase |
| R06437 | dTDP-4-dehydro-6-deoxy-alpha-D-glucose 3-epimerase |
| R06438 | dTDP-6-deoxy-alpha-D-allose:NAD+ oxidoreductase |
| R06439 | S-adenosyl-L-methionine:dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose 3-N,N-dimethyltransferase |
| R06513 | dTDPglucose 4,6-hydro-lyase |
| R06514 | dTDP-4-dehydro-6-deoxy-alpha-D-glucose 3,5-epimerase; dTDP-4-dehydro-6-deoxy-D-glucose 3,5-epimerase |
| R06631 | dTDP-3-amino-2,3,6-trideoxy-4-keto-D-glucose:2-oxoglutarate aminotransferase |
| R08583 | dTDP-4-amino-4,6-dideoxy-D-glucose ammonia-lyase (dTDP-3-oxo-4,6-dideoxy-D-glucose-forming) |
| R08597 | dTDP-4-oxo-2,6-dideoxy-D-glucose 3,5-epimerase |
| R08930 | dTDP-4-dehydro-2,6-dideoxy-alpha-D-glucose hydro-lyase (dTDP-2,3,6-trideoxy-alpha-D-hexopyranose-forming) |
| R08931 | dTDP-4-amino-2,3,4,6-tetradeoxy-D-glucose:2-oxoglutarate aminotransferase |
| R08932 | S-adenosyl-L-methionine:dTDP-4-amino-2,3,4,6-tetradeoxy-D-glucose N,N-dimethyltransferase |
| R08933 | S-adenosyl-L-methionine:dTDP-4-oxo-2,6-dideoxy-D-allose 3-C-methyltransferase |
| R08934 | dTDP-3-methyl-4-oxo-2,6-dideoxy-D-glucose 5-epimerase |
| R08935 | dTDP-6-deoxy-L-mannose:NADP+ 4-oxidoreductase (3,5-epimerizing) |
| R09821 | S-adenosyl-L-methionine:dTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose 3-N,N-dimethyltransferase |
| R09824 | dTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose:2-oxoglutarate aminotransferase |
| R09873 | acetyl-CoA:dTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose 3-N-acetyltransferase |
| R09904 | dTDP-4-dehydro-6-deoxy-alpha-D-glucopyranose:dTDP-3-dehydro-6-deoxy-alpha-D-galactopyranose isomerase |
| R10190 | dTDP-6-deoxy-beta-L-talose:NAD+ 4-oxidoreductase |
| R10279 | dTDP-6-deoxy-beta-L-talose 4-epimerase |
| R10459 | dTDP-5-dimethyl-L-lyxose:NAD+ 4-oxidoreductase |
| R10536 | dTDP-alpha-D-fucopyranose furanomutase |
| R10573 | dTDP-6-deoxy-alpha-D-allose:NADP+ oxidoreductase |
| R11020 | S-adenosyl-L-methionine:dTDP-4-oxo-6-deoxy-D-allose 5-C-methyltransferase |
| R11041 | S-adenosyl-L-methionine:dTDP-4-oxo-2,6-dideoxy-L-mannose 3-O-methyltransferase |
| R11045 | S-adenosyl-L-methionine:dTDP-3-amino-2,3,6-trideoxy-4-keto-D-glucose 3-N,N-dimethyltransferase |
| R11046 | dTDP-3-N,N-dimethylamino-2,3,6-trideoxy-4-keto-D-glucose 5-epimerase |
| R11051 | dTDP-4-oxo-2,6-dideoxy-D-allose:NADP+ 3-oxidoreductase |
| R11468 | dTDP-4-amino-4,6-dideoxy-D-glucose:reduced-ferredoxin:oxygen oxidoreductase |
| R11469 | S-adenosyl-L-methionine:dTDP-L-rhamnose 3'-O-methyltransferase |
| R11471 | dTDP-glucose:NAD+ 6-oxidoreductase |
| R11472 | dTDP-D-glucuronate:NAD+ oxidoreductase (decarboxylating) |
| R11473 | dTDP-4-oxo-alpha-D-xylose hydro-lyase (dTDP-4-oxo-2-deoxy-alpha-D-pentos-2-ene-forming) |
| R11474 | dTDP-4-oxo-2-deoxy-beta-L-xylose:NADP+ oxidoreductase |
| R11475 | dTDP-4-amino-2,4-dideoxy-beta-L-xylose:2-oxoglutarate aminotransferase |
| R11476 | S-adenosyl-L-methionine:dTDP-4-amino-2,4-dideoxy-beta-L-xylose N-methyltransferase |
|
|---|
| Compound |
| C00688 | dTDP-4-dehydro-beta-L-rhamnose |
| C03187 | dTDP-6-deoxy-beta-L-talose |
| C04268 | dTDP-4-amino-4,6-dideoxy-D-glucose |
| C06620 | dTDP-3,4-dioxo-2,6-dideoxy-D-glucose |
| C11447 | dTDP-4-dimethylamino-4,6-dideoxy-5-C-methyl-D-allose |
| C11460 | dTDP-4-oxo-5-C-methyl-L-rhamnose |
| C11461 | dTDP-4-amino-4,6-dideoxy-5-C-methyl-D-allose |
| C11907 | dTDP-4-oxo-6-deoxy-D-glucose |
| C11908 | dTDP-3-oxo-6-deoxy-D-glucose |
| C11909 | dTDP-3-oxo-4,6-dideoxy-D-glucose |
| C11910 | dTDP-3-amino-3,4,6-trideoxy-D-glucose |
| C11915 | dTDP-3-methyl-4-oxo-2,6-dideoxy-L-allose |
| C11922 | dTDP-4-oxo-2,6-dideoxy-D-glucose |
| C11925 | dTDP-3-amino-3,6-dideoxy-D-glucose |
| C11926 | dTDP-4-oxo-6-deoxy-D-allose |
| C11927 | dTDP-4-oxo-2,6-dideoxy-L-mannose |
| C11928 | dTDP-4-oxo-2,6-dideoxy-D-allose |
| C12318 | dTDP-3-amino-2,3,6-trideoxy-4-keto-D-glucose |
| C12481 | dTDP-5-dimethyl-L-lyxose |
| C18029 | dTDP-3-oxo-2,6-dideoxy-D-glucose |
| C18030 | dTDP-2,6-dideoxy-D-kanosamine |
| C18032 | dTDP-4-oxo-2,3,6-trideoxy-D-glucose |
| C18033 | dTDP-4-amino-2,3,4,6-tetradeoxy-D-glucose |
| C18035 | dTDP-3-methyl-4-oxo-2,6-dideoxy-D-glucose |
| C19947 | dTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose |
| C19948 | dTDP-3-dimethylamino-3,6-dideoxy-alpha-D-galactopyranose |
| C19960 | dTDP-3-dehydro-6-deoxy-alpha-D-galactopyranose |
| C20156 | dTDP-3-acetamido-3,6-dideoxy-alpha-D-galactopyranose |
| C20682 | dTDP-alpha-D-fucofuranose |
| C21071 | dTDP-3-O-methyl-4-oxo-2,6-dideoxy-L-mannose |
| C21073 | dTDP-3-N,N-dimethylamino-2,3,6-trideoxy-4-keto-D-glucose |
| C21074 | dTDP-3-N,N-dimethylamino-4-oxo-2,3,6-trideoxy-L-allose |
| C21348 | dTDP-4-hydroxyamino-4,6-dideoxy-alpha-D-glucose |
| C21349 | dTDP-4-deoxy-4-thio-alpha-D-digitoxose |
| C21350 | dTDP-3-O-methyl-beta-L-rhamnose |
| C21351 | dTDP-4-oxo-alpha-D-xylose |
| C21352 | dTDP-4-oxo-2-deoxy-alpha-D-pentos-2-ene |
| C21353 | dTDP-4-oxo-2-deoxy-beta-L-xylose |
| C21354 | dTDP-4-amino-2,4-dideoxy-beta-L-xylose |
| C21355 | dTDP-4-methylamino-2,4-dideoxy-beta-L-xylose |
| C21356 | dTDP-4-ethylamino-3-O-methyl-2,4-dideoxy-L-threo-pentopyranose |
| C21362 | dTDP-3-methyl-4-oxo-alpha-D-ribopyranose |
|
|---|
| Reference |
|
|---|
| Authors |
Rawlings BJ. |
|---|
| Title |
Type I polyketide biosynthesis in bacteria (Part A--erythromycin biosynthesis). |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rawlings BJ. |
|---|
| Title |
Type I polyketide biosynthesis in bacteria (part B). |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wohlert S, Lomovskaya N, Kulowski K, Fonstein L, Occi JL, Gewain KM, MacNeil DJ, Hutchinson CR |
|---|
| Title |
Insights about the biosynthesis of the avermectin deoxysugar L-oleandrose through heterologous expression of Streptomyces avermitilis deoxysugar genes in Streptomyces lividans. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Bate N, Butler AR, Smith IP, Cundliffe E |
|---|
| Title |
The mycarose-biosynthetic genes of Streptomyces fradiae, producer of tylosin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Amann S, Drager G, Rupprath C, Kirschning A, Elling L |
|---|
| Title |
(Chemo)enzymatic synthesis of dTDP-activated 2,6-dideoxysugars as building blocks of polyketide antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Thibodeaux CJ, Melancon CE, Liu HW |
|---|
| Title |
Unusual sugar biosynthesis and natural product glycodiversification. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Volchegursky Y, Hu Z, Katz L, McDaniel R |
|---|
| Title |
Biosynthesis of the anti-parasitic agent megalomicin: transformation of erythromycin to megalomicin in Saccharopolyspora erythraea. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mironov VA, Sergienko OV, Nastasiak IN, Danilenko VN. |
|---|
| Title |
Biogenesis and regulation of biosynthesis of erythromycins in Saccharopolyspora. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Tello M, Jakimowicz P, Errey JC, Freel Meyers CL, Walsh CT, Buttner MJ, Lawson DM, Field RA |
|---|
| Title |
Characterisation of Streptomyces spheroides NovW and revision of its functional assignment to a dTDP-6-deoxy-D-xylo-4-hexulose 3-epimerase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Takahashi H, Liu YN, Chen H, Liu HW |
|---|
| Title |
Biosynthesis of TDP-l-mycarose: the specificity of a single enzyme governs the outcome of the pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Brana AF, Mendez C, Salas JA |
|---|
| Title |
Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rodriguez L, Rodriguez D, Olano C, Brana AF, Mendez C, Salas JA |
|---|
| Title |
Functional analysis of OleY L-oleandrosyl 3-O-methyltransferase of the oleandomycin biosynthetic pathway in Streptomyces antibioticus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Useglio M, Peiru S, Rodriguez E, Labadie GR, Carney JR, Gramajo H |
|---|
| Title |
TDP-L-megosamine biosynthesis pathway elucidation and megalomicin a production in Escherichia coli. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Liu W, Christenson SD, Standage S, Shen B |
|---|
| Title |
Biosynthesis of the enediyne antitumor antibiotic C-1027. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B |
|---|
| Title |
Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Gao Q, Zhang C, Blanchard S, Thorson JS |
|---|
| Title |
Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: insights from the AT2433 biosynthetic locus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Simkhada D, Oh TJ, Kim EM, Yoo JC, Sohng JK |
|---|
| Title |
Cloning and characterization of CalS7 from Micromonospora echinospora sp. calichensis as a glucose-1-phosphate nucleotidyltransferase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Singh S, Peltier-Pain P, Tonelli M, Thorson JS |
|---|
| Title |
A general NMR-based strategy for the in situ characterization of sugar-nucleotide-dependent biosynthetic pathways. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Singh S, Chang A, Helmich KE, Bingman CA, Wrobel RL, Beebe ET, Makino S, Aceti DJ, Dyer K, Hura GL, Sunkara M, Morris AJ, Phillips GN Jr, Thorson JS |
|---|
| Title |
Structural and functional characterization of CalS11, a TDP-rhamnose 3'-O-methyltransferase involved in calicheamicin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Liang ZX |
|---|
| Title |
Complexity and simplicity in the biosynthesis of enediyne natural products. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Singh S, Michalska K, Bigelow L, Endres M, Kharel MK, Babnigg G, Yennamalli RM, Bingman CA, Joachimiak A, Thorson JS, Phillips GN Jr |
|---|
| Title |
Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Peltier-Pain P, Singh S, Thorson JS |
|---|
| Title |
Characterization of Early Enzymes Involved in TDP-Aminodideoxypentose Biosynthesis en Route to Indolocarbazole AT2433. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Singh S, Kim Y, Wang F, Bigelow L, Endres M, Kharel MK, Babnigg G, Bingman CA, Joachimiak A, Thorson JS, Phillips GN Jr |
|---|
| Title |
Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00520 | Amino sugar and nucleotide sugar metabolism |
| rn00522 | Biosynthesis of 12-, 14- and 16-membered macrolides |
| rn01059 | Biosynthesis of enediyne antibiotics |
|
|---|
| KO pathway |
|
|---|