| Entry |
|
|---|
| Name |
Biosynthesis of type II polyketide products |
|---|
| Class |
Metabolism; Metabolism of terpenoids and polyketides
|
|---|
| Pathway map |
| rn01057 | Biosynthesis of type II polyketide products |
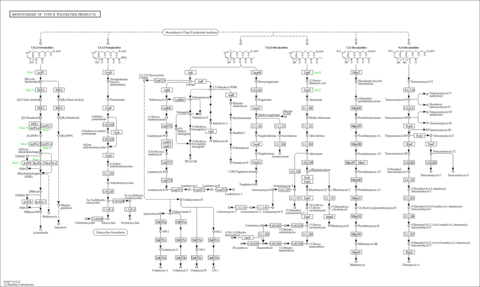
|
|---|
| Module |
| M00779 | Dihydrokalafungin biosynthesis, octaketide => dihydrokalafungin [PATH:rn01057] |
| M00780 | Tetracycline/oxytetracycline biosynthesis, pretetramide => tetracycline/oxytetracycline [PATH:rn01057] |
| M00781 | Nogalavinone/aklavinone biosynthesis, deoxynogalonate/deoxyaklanonate => nogalavinone/aklavinone [PATH:rn01057] |
| M00782 | Mithramycin biosynthesis, 4-demethylpremithramycinone => mithramycin [PATH:rn01057] |
| M00783 | Tetracenomycin C/8-demethyltetracenomycin C biosynthesis, tetracenomycin F2 => tetracenomycin C/8-demethyltetracenomycin C [PATH:rn01057] |
| M00784 | Elloramycin biosynthesis, 8-demethyltetracenomycin C => elloramycin A [PATH:rn01057] |
| M00823 | Chlortetracycline biosynthesis, pretetramide => chlortetracycline [PATH:rn01057] |
|
|---|
| Drug |
| D00201 | Tetracycline (JAN/USP/INN) |
| D07689 | Chlortetracycline (INN) |
|
|---|
| Reaction |
| R04060 | anhydrotetracycline,NADPH:oxygen oxidoreductase (6-hydroxylating) |
| R05459 | tetracycline:coenzyme-F420 oxidoreductase |
| R05462 | 4-hydroxy-6-methylpretetramide,NADPH:oxygen oxidoreductase (12a-hydroxylating) |
| R05463 | 6-methylpretetramide,NADPH:oxygen oxidoreductase (4-hydroxylating) |
| R05705 | FMNH2:NAD+ oxidoreductase |
| R06660 | dTDP-D-olivose:compound-100-1 O-beta-D-olivosyltransferase |
| R06668 | dTDP-D-olivose:urdamycin-G D-olivosyltransferase |
| R06675 | S-adenosyl-L-methionine:nogalonic-acid O-methyltransferase |
| R06676 | auraviketone lyase (decyclizing) |
| R06677 | auramycinone:NADP+ 7-oxidoreductase |
| R06679 | 12-deoxyaklanonic-acid:oxygen oxidoreductase |
| R06680 | S-adenosyl-L-methionine:aklanonate O-methyltransferase |
| R06681 | aklaviketone lyase (ring-opening); methyl aklanonate cyclase |
| R06682 | aklavinone:NADP+ 7-oxidoreductase |
| R06683 | aklavinone,NADPH:oxygen oxidoreductase (12-hydroxylating) |
| R06684 | dTDP-beta-L-daunosamine:epsilon-rhodomycinone beta-L-daunosaminyltransferase |
| R06687 | S-adenosyl-L-methionine:13-deoxycarminomycin 4-O-methyltransferase |
| R06688 | 13-deoxydaunorubicin,NADPH:oxygen oxidoreductase (13-hydroxylating) |
| R06689 | 13-dihydrodaunorubicin,NADPH:oxygen oxidoreductase (13-hydroxylating) |
| R06690 | daunorubicin,NADPH:oxygen oxidoreductase (hydroxylating) |
| R06691 | 13-deoxycarminomycin,NADPH:oxygen oxidoreductase (13-hydroxylating) |
| R06692 | 13-dihydrocarminomycin,NADPH:oxygen oxidoreductase |
| R06693 | S-adenosyl-L-methionine:carminomycin 4-O-methyltransferase |
| R06699 | tetracenomycin-F2 hydro-lyase (tetracenomycin-F1-forming) |
| R06700 | tetracenomycin-F1:oxygen C5-monooxygenase |
| R06703 | S-adenosyl-L-methionine:tetracenomycin-E O-methyltransferase |
| R06704 | tetracenomycin-A2,NADPH:oxygen oxidoreductase (tetracenomycin-C-forming) |
| R06713 | S-adenosyl-L-methionine:tetracenomycin-B3 O-methyltransferase |
| R06715 | S-adenosyl-L-methionine:4-demethylpremithramycinone O-methyltransferase |
| R06716 | dTDP-D-olivose:premithramycinone D-olivosyltransferase |
| R06717 | dTDP-D-oliose:premithramycin-A1 D-oliosyltransferase |
| R06719 | S-adenosyl-L-methionine:premithramycin-A3' C-methyltransferase |
| R09189 | S-adenosyl-L-methionine:pretetramid C-methyltransferase |
| R09190 | 6-methylpretetramide,NADPH:oxygen oxidoreductase (4,12a-hydroxylating) |
| R09191 | S-adenosyl-L-methionine:4-amino-anhydrotetracycline N-methyltransferase |
| R09192 | oxytetracycline:coenzyme-F420 oxidoreductase |
| R09198 | 5a,11a-dehydrotetracycline,NADPH:oxygen oxidoreductase (5-hydroxylating) |
| R09313 | dihydrokalafungin-dihydroquinone,FMNH2:oxygen oxidoreductase (hydroxylating) |
| R09327 | deoxynogalonate:oxygen oxidoreductase |
| R09328 | nogalaviketone lyase (ring-opening); methyl nogalonate cyclase |
| R09329 | nogalavinone:NADP+ 7-oxidoreductase |
| R09331 | dTDP-beta-L-rhodosamine:aklavinone 7-alpha-L-rhodosaminyltransferase |
| R09332 | dTDP-2-deoxy-beta-L-fucose:aclacinomycin-T 2-deoxy-alpha-L-fucosyltransferase |
| R09334 | aclacinomycin N:oxygen oxidoreductase |
| R09335 | aclacinomycin A:oxygen oxidoreductase |
| R09337 | epsilon-rhodomycin-T acylhydrolase |
| R09345 | dTDP-D-olivose:UWM6 9-C-beta-D-olivosyltransferase |
| R09351 | dTDP-D-olivose:premithramycin-A3 D-olivosyltransferase |
| R09352 | dTDP-D-olivose:3A-deolivosylpremithramycin-B D-olivosyltransferase |
| R09353 | premithramycin-B,NADPH:oxygen oxidoreductase |
| R10545 | aclacinomycin T acylhydrolase |
| R10954 | tetracenomycin-B2,NADPH:oxygen oxidoreductase (8-demethyltetracenomycin-C-forming) |
| R10955 | dTDP-beta-L-rhamnose:8-demethyltetracenomycin-C alpha-L-rhamnosyltransferase |
| R10956 | S-adenosyl-L-methionine:8-demethyl-8-alpha-L-rhamnosyltetracenomycin-C 2'-O-methyltransferase |
| R10957 | S-adenosyl-L-methionine:8-demethyl-8-(2-methoxy-alpha-L-rhamnosyl)tetracenomycin-C 3'-O-methyltransferase |
| R10958 | S-adenosyl-L-methionine:8-demethyl-8-(2,3-di-O-methoxy-alpha-L-rhamnosyl)tetracenomycin-C 4'-O-methyltransferase |
| R10959 | S-adenosyl-L-methionine:8-demethyl-8-(2,3,4-tri-O-methyl-alpha-L-rhamnosyl)tetracenomycin-C O-methyltransferase |
| R11478 | tetracycline:FADH2 oxidoreductase (7-halogenating) |
|
|---|
| Compound |
| C03206 | 5a,11a-Dehydrotetracycline |
| C06627 | 4-Keto-anhydrotetracycline |
| C06628 | 4-Hydroxy-6-methylpretetramide |
| C06654 | 4-Amino-anhydrotetracycline |
| C12373 | Tetracenomycin F1 methylester |
| C12374 | Decarboxytetracenomycin F1 |
| C12375 | Tetracenomycin D3 methylester |
| C12379 | 8-Demethyltetracenomycin C |
| C12382 | 4-Demethylpremithramycinone |
| C12400 | 19-Hydroxy-8-O-methyltetrangulol |
| C12427 | 10-Carboxy-13-deoxycarminomycin |
| C12433 | (13S)-13-Dihydrodaunorubicin |
| C12435 | 6-Deoxydihydrokalafungin |
| C18293 | Nonaketamide monocyclic intermediate |
| C18294 | Nonaketamide tricyclic intermediate |
| C18296 | 5a,11a-Dehydrooxytetracycline |
| C18329 | 7,9,12-Octaketide intermediate 3 |
| C18332 | 7,9,12-Decaketide intermediate 3 |
| C18335 | 7,9,12-Decaketide intermediate 6 |
| C18336 | 7,12-Decaketide intermediate 1 |
| C18337 | 9,14-Decaketide intermediate 1 |
| C18353 | Octaketide bicyclic intermediate |
| C18360 | Dihydrokalafungin dihydroquinone form |
| C18641 | 15-Demethoxy-epsilon-rhodomycin |
| C18681 | C1'-C9-Glycosylated UWM6 |
| C18709 | Decaketide tricyclic intermediate |
| C18710 | 3A-Deolivosylpremithramycin B |
| C18778 | 8-D-Olivosyl-landomycin |
| C20689 | 15-Demethylaclacinomycin T |
| C20974 | 8-Demethyl-8-alpha-L-rhamnosyltetracenomycin C |
| C20975 | 8-Demethyl-8-(2-O-methyl-alpha-L-rhamnosyl)tetracenomycin C |
| C20976 | 8-Demethyl-8-(2,3-di-O-methyl-alpha-L-rhamnosyl)tetracenomycin C |
| C20977 | 8-Demethyl-8-(2,3,4-tri-O-methyl-alpha-L-rhamnosyl)tetracenomycin C |
|
|---|
| Reference |
|
|---|
| Authors |
Taguchi T, Okamoto S, Lezhava A, Li A, Ochi K, Ebizuka Y, Ichinose K |
|---|
| Title |
Possible involvement of ActVI-ORFA in transcriptional regulation of actVI tailoring-step genes for actinorhodin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y |
|---|
| Title |
Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Li A, Itoh T, Taguchi T, Xiang T, Ebizuka Y, Ichinose K |
|---|
| Title |
Functional studies on a ketoreductase gene from Streptomyces sp. AM-7161 to control the stereochemistry in medermycin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ichinose K, Bedford DJ, Tornus D, Bechthold A, Bibb MJ, Revill WP, Floss HG, Hopwood DA |
|---|
| Title |
The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tu22: sequence analysis and expression in a heterologous host. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hopwood DA |
|---|
| Title |
Genetic Contributions to Understanding Polyketide Synthases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Fernandez-Moreno MA, Martinez E, Caballero JL, Ichinose K, Hopwood DA, Malpartida F |
|---|
| Title |
DNA sequence and functions of the actVI region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor A3(2). |
|---|
| Journal |
J Biol Chem 269:24854-63 (1994) |
|---|
| Reference |
|
|---|
| Authors |
Itoh T, Taguchi T, Kimberley MR, Booker-Milburn KI, Stephenson GR, Ebizuka Y, Ichinose K |
|---|
| Title |
Actinorhodin biosynthesis: structural requirements for post-PKS tailoring intermediates revealed by functional analysis of ActVI-ORF1 reductase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ichinose K, Taguchi T, Bedford DJ, Ebizuka Y, Hopwood DA |
|---|
| Title |
Functional complementation of pyran ring formation in actinorhodin biosynthesis in Streptomyces coelicolor A3(2) by ketoreductase genes for granaticin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Taguchi T, Ebizuka Y, Hopwood DA, Ichinose K |
|---|
| Title |
A new mode of stereochemical control revealed by analysis of the biosynthesis of dihydrogranaticin in Streptomyces violaceoruber Tu22. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Taguchi T, Kunieda K, Takeda-Shitaka M, Takaya D, Kawano N, Kimberley MR, Booker-Milburn KI, Stephenson GR, Umeyama H, Ebizuka Y, Ichinose K |
|---|
| Title |
Remarkably different structures and reaction mechanisms of ketoreductases for the opposite stereochemical control in the biosynthesis of BIQ antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Okamoto S, Taguchi T, Ochi K, Ichinose K |
|---|
| Title |
Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Valton J, Mathevon C, Fontecave M, Niviere V, Ballou DP |
|---|
| Title |
Mechanism and regulation of the Two-component FMN-dependent monooxygenase ActVA-ActVB from Streptomyces coelicolor. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Valton J, Fontecave M, Douki T, Kendrew SG, Niviere V |
|---|
| Title |
An aromatic hydroxylation reaction catalyzed by a two-component FMN-dependent Monooxygenase. The ActVA-ActVB system from Streptomyces coelicolor. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Valton J, Filisetti L, Fontecave M, Niviere V |
|---|
| Title |
A two-component flavin-dependent monooxygenase involved in actinorhodin biosynthesis in Streptomyces coelicolor. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ellis HR |
|---|
| Title |
The FMN-dependent two-component monooxygenase systems. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Filisetti L, Fontecave M, Niviere V |
|---|
| Title |
Mechanism and substrate specificity of the flavin reductase ActVB from Streptomyces coelicolor. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kendrew SG, Hopwood DA, Marsh EN |
|---|
| Title |
Identification of a monooxygenase from Streptomyces coelicolor A3(2) involved in biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sciara G, Kendrew SG, Miele AE, Marsh NG, Federici L, Malatesta F, Schimperna G, Savino C, Vallone B |
|---|
| Title |
The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Fetzner S, Steiner RA |
|---|
| Title |
Cofactor-independent oxidases and oxygenases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Krohn K (ed). |
|---|
| Title |
Anthracycline Chemistry and Biology I : Biological Occurence and Biosynthesis, Synthesis and Chemistry |
|---|
| Journal |
Topics in Current Chemistry 282 (2008) |
|---|
| Reference |
|
|---|
| Authors |
Torkkell S, Kunnari T, Palmu K, Mantsala P, Hakala J, Ylihonko K |
|---|
| Title |
The entire nogalamycin biosynthetic gene cluster of Streptomyces nogalater: characterization of a 20-kb DNA region and generation of hybrid structures. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Grocholski T, Koskiniemi H, Lindqvist Y, Mantsala P, Niemi J, Schneider G |
|---|
| Title |
Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Koskiniemi H, Grocholski T, Schneider G, Niemi J |
|---|
| Title |
Expression, purification and crystallization of the cofactor-independent monooxygenase SnoaB from the nogalamycin biosynthetic pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chung JY, Fujii I, Harada S, Sankawa U, Ebizuka Y |
|---|
| Title |
Expression, purification, and characterization of AknX anthrone oxygenase, which is involved in aklavinone biosynthesis in Streptomyces galilaeus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Dickens ML, Ye J, Strohl WR |
|---|
| Title |
Analysis of clustered genes encoding both early and late steps in daunomycin biosynthesis by Streptomyces sp. strain C5. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Torkkell S, Kunnari T, Palmu K, Hakala J, Mantsala P, Ylihonko K |
|---|
| Title |
Identification of a cyclase gene dictating the C-9 stereochemistry of anthracyclines from Streptomyces nogalater. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sultana A, Kallio P, Jansson A, Wang JS, Niemi J, Mantsala P, Schneider G |
|---|
| Title |
Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kallio P, Sultana A, Niemi J, Mantsala P, Schneider G |
|---|
| Title |
Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Dickens ML, Ye J, Strohl WR |
|---|
| Title |
Cloning, sequencing, and analysis of aklaviketone reductase from Streptomyces sp. strain C5. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Leimkuhler C, Fridman M, Lupoli T, Walker S, Walsh CT, Kahne D |
|---|
| Title |
Characterization of rhodosaminyl transfer by the AknS/AknT glycosylation complex and its use in reconstituting the biosynthetic pathway of aclacinomycin A. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lu W, Leimkuhler C, Gatto GJ Jr, Kruger RG, Oberthur M, Kahne D, Walsh CT |
|---|
| Title |
AknT is an activating protein for the glycosyltransferase AknS in L-aminodeoxysugar transfer to the aglycone of aclacinomycin A. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lu W, Leimkuhler C, Oberthur M, Kahne D, Walsh CT |
|---|
| Title |
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Alexeev I, Sultana A, Mantsala P, Niemi J, Schneider G |
|---|
| Title |
Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lindqvist Y, Koskiniemi H, Jansson A, Sandalova T, Schnell R, Liu Z, Mantsala P, Niemi J, Schneider G |
|---|
| Title |
Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Malla S, Niraula NP, Singh B, Liou K, Sohng JK |
|---|
| Title |
Limitations in doxorubicin production from Streptomyces peucetius. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Otten SL, Liu X, Ferguson J, Hutchinson CR |
|---|
| Title |
Cloning and characterization of the Streptomyces peucetius dnrQS genes encoding a daunosamine biosynthesis enzyme and a glycosyl transferase involved in daunorubicin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Malla S, Niraula NP, Liou K, Sohng JK |
|---|
| Title |
Enhancement of doxorubicin production by expression of structural sugar biosynthesis and glycosyltransferase genes in Streptomyces peucetius. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Dickens ML, Priestley ND, Strohl WR |
|---|
| Title |
In vivo and in vitro bioconversion of epsilon-rhodomycinone glycoside to doxorubicin: functions of DauP, DauK, and DoxA. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lomovskaya N, Otten SL, Doi-Katayama Y, Fonstein L, Liu XC, Takatsu T, Inventi-Solari A, Filippini S, Torti F, Colombo AL, Hutchinson CR |
|---|
| Title |
Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Jansson A, Koskiniemi H, Erola A, Wang J, Mantsala P, Schneider G, Niemi J |
|---|
| Title |
Aclacinomycin 10-hydroxylase is a novel substrate-assisted hydroxylase requiring S-adenosyl-L-methionine as cofactor. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wohlert SE, Wendt-Pienkowski E, Bao W, Hutchinson CR |
|---|
| Title |
Production of aromatic minimal polyketides by the daunorubicin polyketide synthase genes reveals the incompatibility of the heterologous DpsY and JadI cyclases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhou H, Li Y, Tang Y |
|---|
| Title |
Cyclization of aromatic polyketides from bacteria and fungi. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A |
|---|
| Title |
Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chen Y, Fan K, He Y, Xu X, Peng Y, Yu T, Jia C, Yang K |
|---|
| Title |
Characterization of JadH as an FAD- and NAD(P)H-dependent bifunctional hydroxylase/dehydrase in jadomycin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rix U, Wang C, Chen Y, Lipata FM, Remsing Rix LL, Greenwell LM, Vining LC, Yang K, Rohr J |
|---|
| Title |
The oxidative ring cleavage in jadomycin biosynthesis: a multistep oxygenation cascade in a biosynthetic black box. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chen YH, Wang CC, Greenwell L, Rix U, Hoffmeister D, Vining LC, Rohr J, Yang KQ |
|---|
| Title |
Functional analyses of oxygenases in jadomycin biosynthesis and identification of JadH as a bifunctional oxygenase/dehydrase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Borissow CN, Graham CL, Syvitski RT, Reid TR, Blay J, Jakeman DL |
|---|
| Title |
Stereochemical integrity of oxazolone ring-containing jadomycins. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mittler M, Bechthold A, Schulz GE |
|---|
| Title |
Structure and action of the C-C bond-forming glycosyltransferase UrdGT2 involved in the biosynthesis of the antibiotic urdamycin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Trefzer A, Hoffmeister D, Kunzel E, Stockert S, Weitnauer G, Westrich L, Rix U, Fuchser J, Bindseil KU, Rohr J, Bechthold A |
|---|
| Title |
Function of glycosyltransferase genes involved in urdamycin A biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hoffmeister D, Ichinose K, Bechthold A |
|---|
| Title |
Two sequence elements of glycosyltransferases involved in urdamycin biosynthesis are responsible for substrate specificity and enzymatic activity. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rohr J, Schonewolf M, Udvarnoki G, Eckardt K, Schumann G, Wagner C, Beale JM, Sorey SD. |
|---|
| Title |
Investigations on the biosynthesis of the angucycline group antibiotics aquayamycin and the urdamycins A and B. Results from the structural analysis of novel blocked mutant products. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhu L, Ostash B, Rix U, Nur-E-Alam M, Mayers A, Luzhetskyy A, Mendez C, Salas JA, Bechthold A, Fedorenko V, Rohr J |
|---|
| Title |
Identification of the function of gene lndM2 encoding a bifunctional oxygenase-reductase involved in the biosynthesis of the antitumor antibiotic landomycin E by Streptomyces globisporus 1912 supports the originally assigned structure for landomycinone. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Baig I, Kharel M, Kobylyanskyy A, Zhu L, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko VA, Rohr J |
|---|
| Title |
On the acceptor substrate of C-glycosyltransferase UrdGT2: three prejadomycin C-Glycosides from an engineered mutant of Streptomyces globisporus 1912 DeltalndE(urdGT2). |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ostash B, Rix U, Rix LL, Liu T, Lombo F, Luzhetskyy A, Gromyko O, Wang C, Brana AF, Mendez C, Salas JA, Fedorenko V, Rohr J |
|---|
| Title |
Generation of new landomycins by combinatorial biosynthetic manipulation of the LndGT4 gene of the landomycin E cluster in S. globisporus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lombo F, Menendez N, Salas JA, Mendez C |
|---|
| Title |
The aureolic acid family of antitumor compounds: structure, mode of action, biosynthesis, and novel derivatives. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Trefzer A, Blanco G, Remsing L, Kunzel E, Rix U, Lipata F, Brana AF, Mendez C, Rohr J, Bechthold A, Salas JA |
|---|
| Title |
Rationally designed glycosylated premithramycins: hybrid aromatic polyketides using genes from three different biosynthetic pathways. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lozano MJ, Remsing LL, Quiros LM, Brana AF, Fernandez E, Sanchez C, Mendez C, Rohr J, Salas JA |
|---|
| Title |
Characterization of two polyketide methyltransferases involved in the biosynthesis of the antitumor drug mithramycin by Streptomyces argillaceus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Blanco G, Fernandez E, Fernandez MJ, Brana AF, Weissbach U, Kunzel E, Rohr J, Mendez C, Salas JA |
|---|
| Title |
Characterization of two glycosyltransferases involved in early glycosylation steps during biosynthesis of the antitumor polyketide mithramycin by Streptomyces argillaceus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Nur-e-Alam M, Mendez C, Salas JA, Rohr J |
|---|
| Title |
Elucidation of the glycosylation sequence of mithramycin biosynthesis: isolation of 3A-deolivosylpremithramycin B and its conversion to premithramycin B by glycosyltransferase MtmGII. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Fernandez E, Weissbach U, Sanchez Reillo C, Brana AF, Mendez C, Rohr J, Salas JA |
|---|
| Title |
Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Beam MP, Bosserman MA, Noinaj N, Wehenkel M, Rohr J |
|---|
| Title |
Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway . |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wang C, Gibson M, Rohr J, Oliveira MA |
|---|
| Title |
Crystallization and X-ray diffraction properties of Baeyer-Villiger monooxygenase MtmOIV from the mithramycin biosynthetic pathway in Streptomyces argillaceus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rodriguez D, Quiros LM, Brana AF, Salas JA |
|---|
| Title |
Purification and characterization of a monooxygenase involved in the biosynthetic pathway of the antitumor drug mithramycin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hutchinson CR |
|---|
| Title |
Biosynthetic Studies of Daunorubicin and Tetracenomycin C. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Summers RG, Wendt-Pienkowski E, Motamedi H, Hutchinson CR |
|---|
| Title |
The tcmVI region of the tetracenomycin C biosynthetic gene cluster of Streptomyces glaucescens encodes the tetracenomycin F1 monooxygenase, tetracenomycin F2 cyclase, and, most likely, a second cyclase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Shen B, Hutchinson CR |
|---|
| Title |
Tetracenomycin F2 cyclase: intramolecular aldol condensation in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Shen B, Hutchinson CR |
|---|
| Title |
Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphthacenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Summers RG, Wendt-Pienkowski E, Motamedi H, Hutchinson CR |
|---|
| Title |
Nucleotide sequence of the tcmII-tcmIV region of the tetracenomycin C biosynthetic gene cluster of Streptomyces glaucescens and evidence that the tcmN gene encodes a multifunctional cyclase-dehydratase-O-methyl transferase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC |
|---|
| Title |
Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rafanan ER Jr, Hutchinson CR, Shen B |
|---|
| Title |
Triple hydroxylation of tetracenomycin A2 to tetracenomycin C involving two molecules of O(2) and one molecule of H(2)O. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ramos A, Lombo F, Brana AF, Rohr J, Mendez C, Salas JA |
|---|
| Title |
Biosynthesis of elloramycin in Streptomyces olivaceus requires glycosylation by enzymes encoded outside the aglycon cluster. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Patallo EP, Blanco G, Fischer C, Brana AF, Rohr J, Mendez C, Salas JA |
|---|
| Title |
Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn01056 | Biosynthesis of type II polyketide backbone |
|
|---|
| KO pathway |
|
|---|