About E-zyme
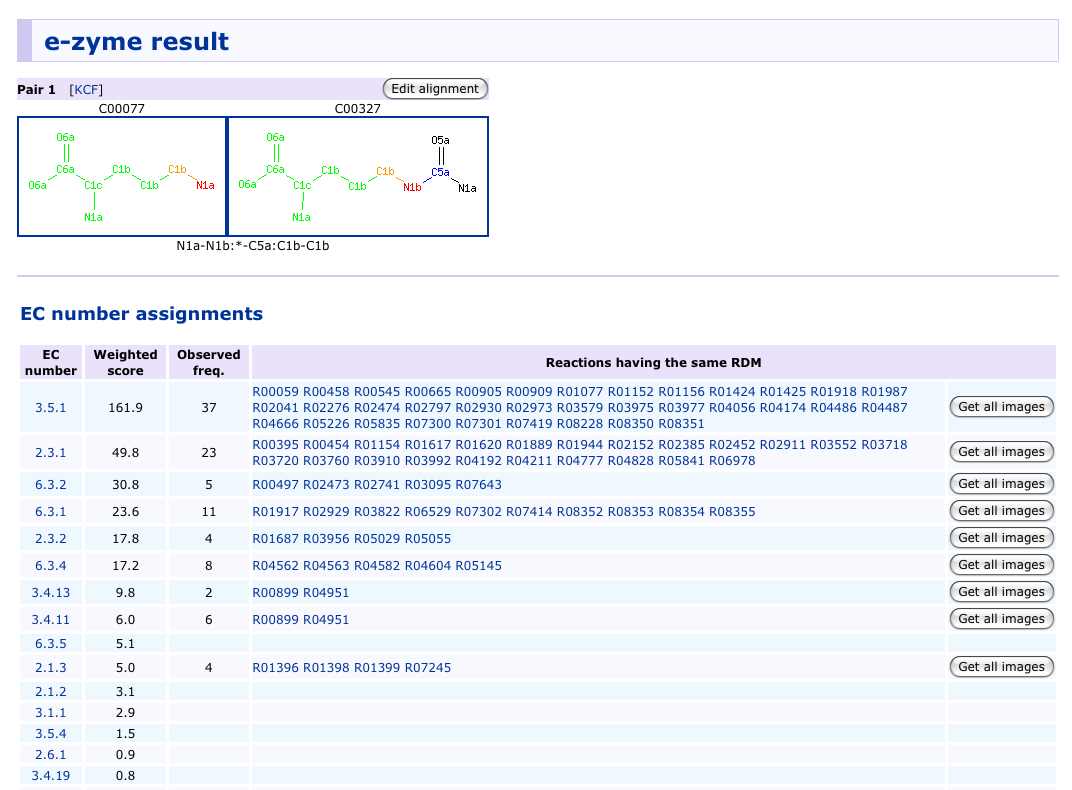
Example of prediction result
|
E-zyme is a computerized method to automatically assign the EC numbers up to the sub-subclasses, i.e., without the fourth serial number for substrate specificity, given pairs of substrates and products. The method is based on a new representation of enzymatic reactions, named the RDM patterns. Each reaction in the current dataset of the EC numbers is first decomposed into reactant pairs. Each pair is then structurally aligned to identify the reaction center (R), the matched region (M), and the difference region (D) by SIMCOMP algorithm. The RDM pattern represents the conversion patterns of atom types in these three regions. (see reference)
RDM patterns
Multi-layered matching and weighted major voting

Reaction pattern profile
|
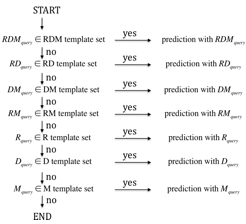
Multi-layered partial matching
|
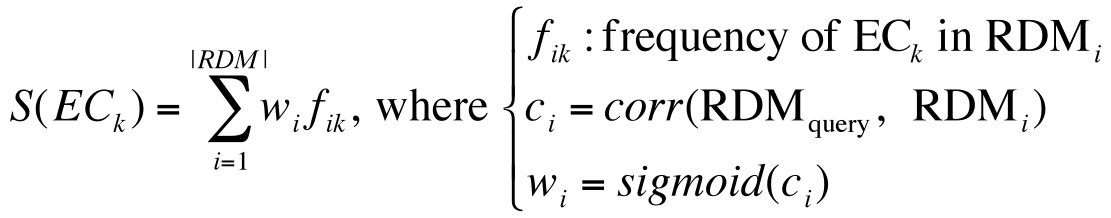
Weighted major voting
|
Prior to the prediction, we prepared a set of template RDM patterns as {RDMi} and a set of EC sub-subclasses as {ECk}. Then, we define a reaction pattern profile for each RDM pattern as {xk}, where the k-th element is the number of reported reactions for the k-th EC sub-subclass. Here, we use the cosine correlation coefficient between reaction pattern profiles xi and xj to evaluate the similarity between them.
To improve the coverage as well as the precision, we use a multi-layered reaction pattern profile scheme. The prediction proceeds according to the following steps. First, the chemical structure alignment outputs the RDM pattern(s) of the query reactant pairs. Consequently, the multi-layered reaction pattern profiles work as sieves: the query that does not match any reaction patterns in the upper layer is passed to the lower layer with looser matching conditions. In each layer a reaction pattern profile is defined to select similar reaction patterns with the query. The layer used to predict EC sub-subclasses will be shown on the bottom of the result page as "Confidence level of prediction results".
In order to select the most appropriate EC sub-subclass out of the candidates, the weighted score is computed for each candidate EC sub-subclass with the equation shown on the right figure. The aim of using the sigmoid function in the weight is to reduce the noise effect of lowly correlated EC sub-subclasses and to put emphasis on highly correlated EC sub-subclasses. After calculation for all EC sub-subclasses in the database, the EC sub-subclass with the highest score is regarded as predicted and will be output on the top of result list.
Other features
- "Multiple pairs" mode is also available via the "Query mode" menu when a user has information of more than one compoud pair, which should be simultaneously involved in a reaciton. When multiple information is given, the prediction accuracy should be improved.
- Links to EC number hierarchy in BRITE database and links to relevant reactions in REACTION database are also provided from result page to consider the appropriateness of the predicted EC sub-subclass.
- A user can edit and manually re-align the atom alignments of each compound pair in order to correct the mis-alignment and/or the mis-assignment of R atoms which will be automatically computed.
References
- Kotera, M., Okuno, Y., Hattori, M., Goto, S., and Kanehisa, M.; Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 126, 16487-16498 (2004). [pubmed]
- Oh, M., Yamada, T., Hattori, M., Goto, S., and Kanehisa, M.; Systematic analysis of enzyme-catalyzed reaction patterns and prediction of microbial biodegradation pathways. J. Chem. Inf. Model. 47, 1702-1712 (2007). [pubmed]
- Yamanishi, Y., Hattori, M., Kotera, M., Goto, S., and Kanehisa, M.; E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics 25, i179-i186 (2009). [pubmed]
Last updated: December 17, 2015
|
|

