"
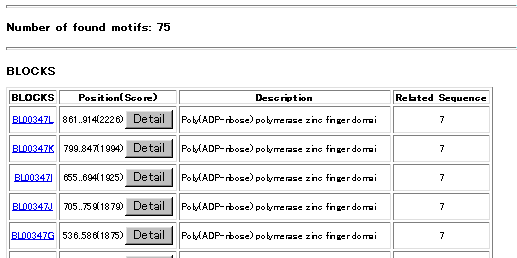
The blocks for the BLOCKS Database are made automatically by looking for the most highly
conserved regions in groups of proteins documented in the Prosite Database. The Prosite pattern
for a protein group is not used in any way to make the BLOCKS Database and the pattern may or
may not be contained in one of the blocks representing a group. These blocks are then calibrated
against the SWISS-PROT database to obtain a measure of the chance distribution of matches. It
is these calibrated blocks that make up the BLOCKS Database."
(From the description of
http://blocks.fhcrc.org)