The PRINTS [ 1, 2 ] is a compendium of protein motif fingerprints. A fingerprint is a group of conserved motifs used to characterise a protein family. It is derived by the excision of conserved motifs from sequence alignments and refined by iterative dredging of the OWL,a non-redundand composite sequence database. Two types of fingerprint are represented in the database, i.e. they are either simple or composite, depending on their complexity: simple fingerprints are essentially single-motifs; while composite fingerprints encode multiple motifs.
This server uses the BLIMPS [3, 4] program to search a protein sequence against the PRINTS database. BLIMPS (BLocks IMProved Searcher) is a searching tool that scores a sequence against blocks or a block against sequences. Original PRINTS database was convered to BLOCKS format for each release of PRINTS. "Composite" fingerprints were decomposed into motif components under the unique PRINTS code (such as "GLABLOOD" held in gc line which is equivalent of "PR00001" in gx line in the PRINTS database.)
During the search each entry in the database, a set of blocks (simple fingerprints or the components of composite ones) is converted to a position-specific score matrix (profile). This matrix is used to score the motif found in the query sequence. The motif blocks in the PRINTS database are calibrated based on the distribution of matches determined in a search as described by the authors. [3] Each raw score is divided by estimated score of 99.5 % of true negative sequence and multiplied by 1000. Thus, a derived score of 1000 means that match would be at the 99.5 % false matches which means scores less than around 1000 are therefore probably not important.
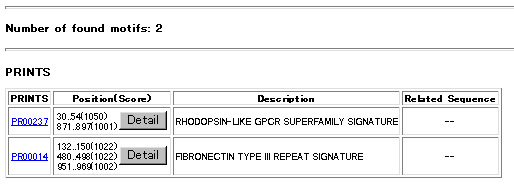
The found motifs are listed in a table. From the gc identifiers of PRINTS you can jump into DBGET to look at the hits as well as related informations precisely. Under Position (Score) column of the table the position (start and end sequence numbers) and the scores of found motifs are listed. Click Detail bottun to see actual positions of the blocks motif along the query sequence . (shown in red)
Below a sample output table of the search is shown. Click the images to get detailed results discribed above.

|