[ GenomeNet Home Page |
Motif Search Home Page |
Motif Help Page ]
Search with a protein sequence against Prodom library
OVERVIEW
The ProDom [1, 2] dabase contains protein
domain families generated from the SWISS-PROT
database by automated sequence comparisons. The current version was built with a new
improved procedure based on recursive PSI-BLAST [3] homology searches.
This server uses the BLAST2 [3] program, the
gapped version of the BLAST to search a protein sequence against the ProDom database.
The cut-off score to evaluate the found motifs used on this server is normalized score or
bit score
expressed in following formula
[3];
S’ = (lam * S - ln K) / ln 2
where S is a raw score of HSP (high-scoreing segment pair) as well as
lam and K are the statistical parameters "lambda" and
"K"
described in the article for BLAST program for statistics.
[4] . This bit score in turn is related to
E value by next formula ;
E = mn / 2 S’
where m and n refer the lengths of the sequences compared. The product mn then represents the
size of search space. Larger bit score threshould gives smaller E value, expected number of HSPs
only by chance matches (false positive), ie., increases the selectivity of the search.
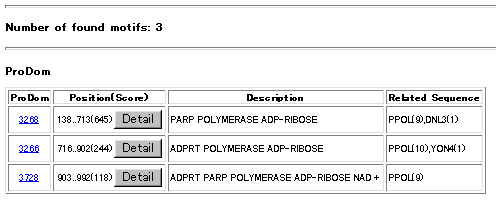
RESULTS
The found motifs are listed in a table. From the ID numbers of ProDom you can jump into DBGET to
look at the hits as well as related informations precisely. Under Position (Score) column of the
table the position (start and end sequence numbers) and
the scores of found motifs are listed. Click Detail bottun to see actua
l positions of the motif along the query sequence .
(shown in red)
OUTPUT
Below a sample output table of the search is shown. Click the images to get detailed results discribed above.
References
-
1.Corpet F., Gouzy J., Kahn D.
Recent improvements of the ProDom
database of protein domain families
Nucleic Acids Res. 27:263-267. (1999)
-
2.
Corpet,F., Gouzy,J. and Kahn,D.
The ProDom database of protein domain families.
Nucleic Acids Res., 26, 323-326 (1998)
-
3.
Altschul,S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,J.L.
Gapped BLAST and
PSI-BLAST: a new generation of protein database search programs
Nucleic Acids Res., 25: 3389-3402.(1997)
-
4.
Karlin S, Altschul SF
Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes.
Proc Natl Acad Sci U S A., 87:2264-8. (1990)