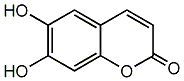
Metabolite
KNApSAcK Entry
| id | C00002471 |
|---|---|
| Name | Esculetin / Aesculetin / 6,7-Dihydroxycoumarin |
| CAS RN | 305-01-1 |
| Standard InChI | InChI=1S/C9H6O4/c10-6-3-5-1-2-9(12)13-8(5)4-7(6)11/h1-4,10-11H |
| Standard InChI (Main Layer) | InChI=1S/C9H6O4/c10-6-3-5-1-2-9(12)13-8(5)4-7(6)11/h1-4,10-11H |
Cluster
| Phytochemical cluster | No. 25 |
|---|---|
| KCF-S cluster | No. 1030 |
Link
ChEMBL
| By standard InChI | CHEMBL244743 |
|---|---|
| By standard InChI Main Layer | CHEMBL244743 |
KEGG
| By LinkDB | C09263 |
|---|
CTD
| By CAS RN | C007628 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 7 |
| asterids | 6 |
| eudicotyledons | 1 |
| Liliopsida | 1 |
Family
| family name | count |
|---|---|
| Asteraceae | 3 |
| Euphorbiaceae | 2 |
| Meliaceae | 2 |
| Lamiaceae | 2 |
| Rutaceae | 2 |
| Plumbaginaceae | 1 |
| Oleaceae | 1 |
| Hippocastanaceae | 1 |
| Xanthorrhoeaceae | 1 |
List (15)
* NCBI
Human Protein / Gene in interaction
40 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | CHEMBL244743 |
CHEMBL1741321
(1)
|
1 / 0 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | CHEMBL244743 |
CHEMBL1908081
(1)
CHEMBL1908082
(1)
|
3 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | CHEMBL244743 |
CHEMBL1613842
(1)
|
4 / 2 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | CHEMBL244743 |
CHEMBL1908087
(1)
|
0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | CHEMBL244743 |
CHEMBL2114784
(2)
|
1 / 1 |
| P04062 | Glucosylceramidase | Enzyme | CHEMBL244743 |
CHEMBL1613818
(1)
|
6 / 4 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | CHEMBL244743 |
CHEMBL1614076
(1)
CHEMBL1614103
(1)
CHEMBL1614031 (1) |
1 / 1 |
| P29466 | Caspase-1 | C14 | CHEMBL244743 |
CHEMBL1614158
(2)
|
0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | CHEMBL244743 |
CHEMBL1614544
(1)
|
11 / 10 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | CHEMBL244743 |
CHEMBL892636
(1)
|
1 / 1 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | CHEMBL244743 |
CHEMBL1741325
(1)
|
0 / 1 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | CHEMBL244743 |
CHEMBL1614166
(1)
|
1 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | CHEMBL244743 |
CHEMBL1008496
(1)
|
0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | CHEMBL244743 |
CHEMBL1614458
(3)
|
0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | CHEMBL244743 |
CHEMBL1614456
(1)
CHEMBL1613803
(1)
|
0 / 0 |
| O75496 | Geminin | Unclassified protein | CHEMBL244743 |
CHEMBL2114843
(1)
CHEMBL2114780
(1)
|
0 / 0 |
| P54855 | UDP-glucuronosyltransferase 2B15 | Enzyme | CHEMBL244743 |
CHEMBL1908090
(1)
|
0 / 0 |
| P15121 | Aldose reductase | Enzyme | CHEMBL244743 |
CHEMBL1260797
(1)
|
0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | CHEMBL244743 |
CHEMBL1963893
(1)
|
0 / 0 |
| P06280 | Alpha-galactosidase A | Enzyme | CHEMBL244743 |
CHEMBL1614217
(3)
CHEMBL1614369
(3)
|
1 / 1 |
| P56817 | Beta-secretase 1 | A1A | CHEMBL244743 |
CHEMBL1936881
(1)
CHEMBL1936882
(1)
|
0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | CHEMBL244743 |
CHEMBL1741322
(1)
|
0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | CHEMBL244743 |
CHEMBL1613910
(2)
CHEMBL1614227
(1)
|
3 / 3 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | CHEMBL244743 |
CHEMBL1614038
(3)
|
2 / 2 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | CHEMBL244743 |
CHEMBL1908085
(1)
|
0 / 0 |
| Q00796 | Sorbitol dehydrogenase | Enzyme | CHEMBL244743 |
CHEMBL1260799
(1)
|
0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | CHEMBL244743 |
CHEMBL1614240
(1)
|
0 / 0 |
| P55210 | Caspase-7 | C14 | CHEMBL244743 |
CHEMBL1613779
(2)
|
0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | CHEMBL244743 |
CHEMBL1741323
(1)
|
1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL244743 |
CHEMBL1614108
(1)
CHEMBL1613886
(1)
CHEMBL1741324 (1) |
0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | CHEMBL244743 |
CHEMBL1794483
(1)
|
0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | CHEMBL244743 |
CHEMBL1737991
(1)
|
0 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | CHEMBL244743 |
CHEMBL1908080
(1)
|
5 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | CHEMBL244743 |
CHEMBL1614421
(1)
|
4 / 3 |
| P34949 | Mannose-6-phosphate isomerase | Enzyme | CHEMBL244743 |
CHEMBL1614255
(1)
CHEMBL1614547
(1)
|
1 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | CHEMBL244743 |
CHEMBL1794536
(2)
|
0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | CHEMBL244743 |
CHEMBL1613914
(3)
|
0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | CHEMBL244743 |
CHEMBL1738442
(1)
|
0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | CHEMBL244743 |
CHEMBL2354311
(1)
|
1 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | CHEMBL244743 |
CHEMBL2114738
(1)
|
0 / 0 |
CTD interaction (28)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| C007628 | 581 |
BAX
BCL2L4 |
BCL2-associated X protein | esculetin affects the localization of BAX protein |
affects localization
|
protein |
19786087
|
| C007628 | 581 |
BAX
BCL2L4 |
BCL2-associated X protein | [esculetin co-treated with Paclitaxel] results in increased expression of BAX protein |
affects cotreatment
/ increases expression |
protein |
16051289
|
| C007628 | 598 |
BCL2L1
BCL-XL/S BCL2L BCLX BCLXL BCLXS Bcl-X PPP1R52 bcl-xL bcl-xS |
BCL2-like 1 | [esculetin co-treated with Paclitaxel] results in decreased expression of BCL2L1 protein |
affects cotreatment
/ decreases expression |
protein |
16051289
|
| C007628 | 637 |
BID
FP497 |
BH3 interacting domain death agonist | [arsenic trioxide co-treated with esculetin] results in increased cleavage of BID protein |
affects cotreatment
/ increases cleavage |
protein |
19428345
|
| C007628 | 836 |
CASP3
CPP32 CPP32B SCA-1 |
caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) | [esculetin co-treated with Paclitaxel] results in increased cleavage of and results in increased activity of CASP3 protein |
affects cotreatment
/ increases activity / increases cleavage |
protein |
16051289
|
| C007628 | 841 |
CASP8
ALPS2B CAP4 Casp-8 FLICE MACH MCH5 |
caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) | [esculetin co-treated with Paclitaxel] results in increased cleavage of and results in increased activity of CASP8 protein |
affects cotreatment
/ increases activity / increases cleavage |
protein |
16051289
|
| C007628 | 1312 |
COMT
|
catechol-O-methyltransferase (EC:2.1.1.6) | COMT protein results in increased methylation of esculetin |
increases methylation
|
protein |
11160877
|
| C007628 | 54205 |
CYCS
CYC HCS THC4 |
cytochrome c, somatic | [arsenic trioxide co-treated with esculetin] affects the localization of CYCS protein |
affects cotreatment
/ affects localization |
protein |
19428345
|
| C007628 | 54205 |
CYCS
CYC HCS THC4 |
cytochrome c, somatic | [esculetin co-treated with Paclitaxel] affects the localization of CYCS protein |
affects cotreatment
/ affects localization |
protein |
16051289
|
| C007628 | 355 |
FAS
ALPS1A APO-1 APT1 CD95 FAS1 FASTM TNFRSF6 |
Fas cell surface death receptor | [esculetin co-treated with Paclitaxel] results in increased expression of FAS protein |
affects cotreatment
/ increases expression |
protein |
16051289
|
| C007628 | 356 |
FASLG
ALPS1B APT1LG1 APTL CD178 CD95-L CD95L FASL TNFSF6 |
Fas ligand (TNF superfamily, member 6) | [esculetin co-treated with Paclitaxel] results in increased expression of FASLG protein |
affects cotreatment
/ increases expression |
protein |
16051289
|
| C007628 | 5594 |
MAPK1
ERK ERK2 ERT1 MAPK2 P42MAPK PRKM1 PRKM2 p38 p40 p41 p41mapk |
mitogen-activated protein kinase 1 (EC:2.7.11.24) | [esculetin co-treated with Paclitaxel] results in decreased expression of MAPK1 protein |
affects cotreatment
/ decreases expression |
protein |
16051289
|
| C007628 | 5594 |
MAPK1
ERK ERK2 ERT1 MAPK2 P42MAPK PRKM1 PRKM2 p38 p40 p41 p41mapk |
mitogen-activated protein kinase 1 (EC:2.7.11.24) | esculetin results in decreased phosphorylation of and results in decreased activity of MAPK1 protein |
decreases activity
/ decreases phosphorylation |
protein |
16051289
|
| C007628 | 1432 |
MAPK14
CSBP CSBP1 CSBP2 CSPB1 EXIP Mxi2 PRKM14 PRKM15 RK SAPK2A p38 p38ALPHA |
mitogen-activated protein kinase 14 (EC:2.7.11.24) | [esculetin co-treated with Paclitaxel] results in decreased expression of MAPK14 protein |
affects cotreatment
/ decreases expression |
protein |
16051289
|
| C007628 | 1432 |
MAPK14
CSBP CSBP1 CSBP2 CSPB1 EXIP Mxi2 PRKM14 PRKM15 RK SAPK2A p38 p38ALPHA |
mitogen-activated protein kinase 14 (EC:2.7.11.24) | esculetin results in decreased phosphorylation of and results in decreased activity of MAPK14 protein |
decreases activity
/ decreases phosphorylation |
protein |
16051289
|
| C007628 | 5595 |
MAPK3
ERK-1 ERK1 ERT2 HS44KDAP HUMKER1A P44ERK1 P44MAPK PRKM3 p44-ERK1 p44-MAPK |
mitogen-activated protein kinase 3 (EC:2.7.11.24) | [esculetin co-treated with Paclitaxel] results in decreased expression of MAPK3 protein |
affects cotreatment
/ decreases expression |
protein |
16051289
|
| C007628 | 5595 |
MAPK3
ERK-1 ERK1 ERT2 HS44KDAP HUMKER1A P44ERK1 P44MAPK PRKM3 p44-ERK1 p44-MAPK |
mitogen-activated protein kinase 3 (EC:2.7.11.24) | esculetin results in decreased phosphorylation of and results in decreased activity of MAPK3 protein |
decreases activity
/ decreases phosphorylation |
protein |
16051289
|
| C007628 | 5599 |
MAPK8
JNK JNK-46 JNK1 JNK1A2 JNK21B1/2 PRKM8 SAPK1 SAPK1c |
mitogen-activated protein kinase 8 (EC:2.7.11.24) | [esculetin co-treated with Paclitaxel] results in increased expression of MAPK8 protein |
affects cotreatment
/ increases expression |
protein |
16051289
|
| C007628 | 5599 |
MAPK8
JNK JNK-46 JNK1 JNK1A2 JNK21B1/2 PRKM8 SAPK1 SAPK1c |
mitogen-activated protein kinase 8 (EC:2.7.11.24) | esculetin results in increased expression of MAPK8 protein |
increases expression
|
protein |
16051289
|
| C007628 | 5599 |
MAPK8
JNK JNK-46 JNK1 JNK1A2 JNK21B1/2 PRKM8 SAPK1 SAPK1c |
mitogen-activated protein kinase 8 (EC:2.7.11.24) | esculetin results in increased phosphorylation of MAPK8 protein |
increases phosphorylation
|
protein |
19786087
|
| C007628 | 5601 |
MAPK9
JNK-55 JNK2 JNK2A JNK2ALPHA JNK2B JNK2BETA PRKM9 SAPK SAPK1a p54a p54aSAPK |
mitogen-activated protein kinase 9 (EC:2.7.11.24) | [esculetin co-treated with Paclitaxel] results in increased expression of MAPK9 protein |
affects cotreatment
/ increases expression |
protein |
16051289
|
| C007628 | 5601 |
MAPK9
JNK-55 JNK2 JNK2A JNK2ALPHA JNK2B JNK2BETA PRKM9 SAPK SAPK1a p54a p54aSAPK |
mitogen-activated protein kinase 9 (EC:2.7.11.24) | esculetin results in increased expression of MAPK9 protein |
increases expression
|
protein |
16051289
|
| C007628 | 4780 |
NFE2L2
NRF2 |
nuclear factor, erythroid 2-like 2 | esculetin results in increased localization of NFE2L2 protein |
increases localization
|
protein |
20933534
|
| C007628 | 1728 |
NQO1
DHQU DIA4 DTD NMOR1 NMORI QR1 |
NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) | esculetin results in increased expression of and results in increased activity of NQO1 protein |
increases activity
/ increases expression |
protein |
20933534
|
| C007628 | 1728 |
NQO1
DHQU DIA4 DTD NMOR1 NMORI QR1 |
NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) | esculetin results in increased expression of NQO1 mRNA |
increases expression
|
mRNA |
20933534
|
| C007628 | 8797 |
TNFRSF10A
APO2 CD261 DR4 TRAILR-1 TRAILR1 |
tumor necrosis factor receptor superfamily, member 10a | esculetin results in increased expression of TNFRSF10A protein |
increases expression
|
protein |
19786087
|
| C007628 | 7157 |
TP53
BCC7 LFS1 P53 TRP53 |
tumor protein p53 | esculetin results in increased degradation of TP53 protein |
increases degradation
|
protein |
14634213
|
| C007628 | 7366 |
UGT2B15
HLUG4 UDPGT_2B8 UDPGT2B15 UDPGTH3 UGT2B8 |
UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) | UGT2B15 protein results in increased metabolism of esculetin |
increases metabolic processing
|
protein |
7835232
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (44)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #602579 | Congenital disorder of glycosylation, type ib; cdg1b |
P34949
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #301500 | Fabry disease |
P06280
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #278300 | Xanthinuria, type i |
P47989
|
KEGG DISEASE (32)
| KEGG | disease name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00125 | Fabry disease |
P06280
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00118 | Congenital disorders of glycosylation (CDG) type I |
P34949
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
Q16637 (related) |
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
Diseases related to CTD interactions
4 disease from interactions of metabolites
| MeSH disease | OMIM | compound | disease name | evidence type |
reference
pmid |
|---|---|---|---|---|---|
| D003093 | C007628 | Colitis, Ulcerative |
therapeutic
|
20380826
|
|
| D003967 | C007628 | Diarrhea |
therapeutic
|
20380826
|
|
| D007674 | C007628 | Kidney Diseases |
therapeutic
|
21450970
|
|
| D014693 | C007628 | Ventricular Fibrillation |
therapeutic
|
1632298
|