Securidaca inappendiculata
Species
KNApSAcK Entry
| Organism name | Securidaca inappendiculata |
|---|---|
| Genus | Securidaca |
| Family | Polygalaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Securidaca |
|---|---|
| Linked NCBI taxonomy ID | 4277 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Polygalaceae |
|---|---|
| ID | 4274 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (11)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00037806
|
Securixanthone B
|
No. 3 | No. 15 |

|
||||
|
C00019303
|
Veratrilogenin
/ 1,7-Dihydroxy-3,4-dimethoxyxanthone |
C031188
|
No. 3 | No. 15 |

|
|||
|
C00037805
|
Securixanthone A
|
No. 3 | No. 15 |

|
||||
|
C00036490
|
2-Hydroxy-1,7-dimethoxyxanthone
|
No. 3 | No. 15 |

|
||||
|
C00035189
|
1,3,7-Trihydroxy-2-methoxyxanthone
|
No. 3 | No. 15 |

|
||||
|
C00036394
|
1,7-Dihydroxy-4-methoxyxanthone
|
CHEMBL484030
|
No. 3 | No. 15 |

|
|||
|
C00036379
|
1,3,8-Trihydroxy-2-methoxyxanthone
|
No. 3 | No. 15 |

|
||||
|
C00036377
|
1,3,6-Trihydroxy-2,7-dimethoxyxanthone
|
No. 3 | No. 15 |

|
||||
|
C00002526
|
Prunetol
/ Genistein / Genisteol / Sophoricol / Differenol A / 1,3,7-Trihydroxyxanthone / 5,7,4'-Trihydroxyisoflavone |
CHEMBL44
|
D019833
|
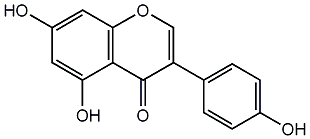
170 / 117 / 106 | 1126 / 38 | No. 71 | No. 15 |
|
|
C00029364
|
Euxanthone
/ 1,7-Dihydroxyxanthone |
CHEMBL389166
|
C404221
|
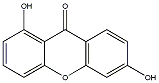
1 / 0 / 0 | No. 76 | No. 15 |

|
|
|
C00031487
|
1,6-Dihydroxyxanthone
|
CHEMBL459695
|
No. 76 | No. 15 |
|
Human Protein / Gene in interactions
171 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00002526 | 1 / 0 |
| P14061 | Estradiol 17-beta-dehydrogenase 1 | Enzyme | C00002526 | 0 / 0 |
| P33527 | Multidrug resistance-associated protein 1 | drugs | C00002526 | 0 / 0 |
| P35610 | Sterol O-acyltransferase 1 | Enzyme | C00029364 | 0 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00002526 | 7 / 37 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00002526 | 4 / 1 |
| Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | Electrochemical transporter | C00002526 | 1 / 0 |
| O94956 | Solute carrier organic anion transporter family member 2B1 | Unclassified protein | C00002526 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002526 | 1 / 1 |
| P14618 | Pyruvate kinase PKM | Enzyme | C00002526 | 0 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00002526 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00002526 | 0 / 3 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00002526 | 2 / 2 |
| P08246 | Neutrophil elastase | S1A | C00002526 | 2 / 1 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00002526 | 0 / 0 |
| Q16539 | Mitogen-activated protein kinase 14 | p38 | C00002526 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00002526 | 0 / 0 |
| P49146 | Neuropeptide Y receptor type 2 | Neuropeptide Y receptor | C00002526 | 0 / 0 |
| P04062 | Glucosylceramidase | Enzyme | C00002526 | 6 / 4 |
| P29466 | Caspase-1 | C14 | C00002526 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00002526 | 0 / 0 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00002526 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00002526 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00002526 | 2 / 2 |
| P02545 | Prelamin-A/C | Unclassified protein | C00002526 | 11 / 10 |
| P00519 | Tyrosine-protein kinase ABL1 | Abl | C00002526 | 1 / 2 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00002526 | 3 / 2 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00002526 | 1 / 1 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00002526 | 3 / 1 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00002526 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00002526 | 1 / 2 |
| P05091 | Aldehyde dehydrogenase, mitochondrial | Oxidoreductase | C00002526 | 1 / 1 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00002526 | 0 / 1 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00002526 | 1 / 1 |
| P25021 | Histamine H2 receptor | Histamine receptor | C00002526 | 0 / 0 |
| P35367 | Histamine H1 receptor | Histamine receptor | C00002526 | 0 / 0 |
| Q01959 | Sodium-dependent dopamine transporter | Dopamine | C00002526 | 1 / 0 |
| P08912 | Muscarinic acetylcholine receptor M5 | Acetylcholine receptor | C00002526 | 0 / 0 |
| P18825 | Alpha-2C adrenergic receptor | Adrenergic receptor | C00002526 | 0 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00002526 | 0 / 0 |
| P11309 | Serine/threonine-protein kinase pim-1 | Pim | C00002526 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00002526 | 1 / 0 |
| P25024 | C-X-C chemokine receptor type 1 | CXC chemokine receptor | C00002526 | 0 / 0 |
| P06241 | Tyrosine-protein kinase Fyn | Src | C00002526 | 0 / 0 |
| Q08209 | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ser_Thr | C00002526 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00002526 | 0 / 1 |
| P14635 | G2/mitotic-specific cyclin-B1 | Other cytosolic protein | C00002526 | 0 / 0 |
| P06493 | Cyclin-dependent kinase 1 | Cdc2 | C00002526 | 0 / 0 |
| P11802 | Cyclin-dependent kinase 4 | CMGC serine/threonine protein kinase family | C00002526 | 1 / 3 |
| P24385 | G1/S-specific cyclin-D1 | Other cytosolic protein | C00002526 | 1 / 8 |
| O75828 | Carbonyl reductase [NADPH] 3 | Enzyme | C00002526 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00002526 | 1 / 2 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00002526 | 1 / 8 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00002526 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00002526 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00002526 | 2 / 3 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | C00002526 | 2 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00002526 | 0 / 0 |
| P37288 | Vasopressin V1a receptor | Vasopressin and oxytocin receptor | C00002526 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00002526 | 0 / 0 |
| Q9Y271 | Cysteinyl leukotriene receptor 1 | Leukotriene receptor | C00002526 | 0 / 0 |
| P29274 | Adenosine receptor A2a | Adenosine receptor | C00002526 | 0 / 0 |
| P11926 | Ornithine decarboxylase | Lyase | C00002526 | 0 / 0 |
| P11474 | Steroid hormone receptor ERR1 | NR3B1 | C00002526 | 0 / 0 |
| P25929 | Neuropeptide Y receptor type 1 | Neuropeptide Y receptor | C00002526 | 0 / 0 |
| Q9Y3R4 | Sialidase-2 | Enzyme | C00002526 | 0 / 0 |
| P50052 | Type-2 angiotensin II receptor | Angiotensin receptor | C00002526 | 1 / 1 |
| P17948 | Vascular endothelial growth factor receptor 1 | Vegfr | C00002526 | 0 / 0 |
| P41968 | Melanocortin receptor 3 | Melanocortin receptor | C00002526 | 1 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002526 | 0 / 0 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | C00002526 | 2 / 2 |
| P04278 | Sex hormone-binding globulin | Secreted protein | C00002526 | 0 / 0 |
| P11509 | Cytochrome P450 2A6 | Cytochrome P450 2A6 | C00002526 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00002526 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002526 | 0 / 0 |
| P16152 | Carbonyl reductase [NADPH] 1 | Enzyme | C00002526 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002526 | 0 / 0 |
| P51151 | Ras-related protein Rab-9A | Unclassified protein | C00002526 | 0 / 0 |
| P38398 | Breast cancer type 1 susceptibility protein | Enzyme | C00002526 | 4 / 2 |
| P04035 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Oxidoreductase | C00002526 | 0 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00002526 | 0 / 0 |
| P21917 | D(4) dopamine receptor | Dopamine receptor | C00002526 | 0 / 0 |
| P30988 | Calcitonin receptor | Calcitonin receptor | C00002526 | 0 / 0 |
| P35462 | D(3) dopamine receptor | Dopamine receptor | C00002526 | 1 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00002526 | 0 / 0 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00002526 | 0 / 1 |
| P41595 | 5-hydroxytryptamine receptor 2B | Serotonin receptor | C00002526 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00002526 | 0 / 0 |
| P25101 | Endothelin-1 receptor | Endothelin receptor | C00002526 | 0 / 0 |
| P30411 | B2 bradykinin receptor | Bradykinin receptor | C00002526 | 0 / 0 |
| P32245 | Melanocortin receptor 4 | Melanocortin receptor | C00002526 | 1 / 0 |
| P32238 | Cholecystokinin receptor type A | Cholecystokinin receptor | C00002526 | 0 / 0 |
| P08311 | Cathepsin G | S1A | C00002526 | 0 / 0 |
| Q99720 | Sigma non-opioid intracellular receptor 1 | Membrane receptor | C00002526 | 1 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00002526 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00002526 | 0 / 1 |
| P32241 | Vasoactive intestinal polypeptide receptor 1 | Vasoactive intestinal peptide receptor | C00002526 | 0 / 0 |
| Q9NPD5 | Solute carrier organic anion transporter family member 1B3 | Electrochemical transporter | C00002526 | 1 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00002526 | 0 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00002526 | 0 / 1 |
| P08172 | Muscarinic acetylcholine receptor M2 | Acetylcholine receptor | C00002526 | 2 / 0 |
| P11229 | Muscarinic acetylcholine receptor M1 | Acetylcholine receptor | C00002526 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00002526 | 2 / 2 |
| P21554 | Cannabinoid receptor 1 | Cannabinoid receptor | C00002526 | 0 / 0 |
| P31645 | Sodium-dependent serotonin transporter | Serotonin | C00002526 | 2 / 0 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00002526 | 5 / 3 |
| P04626 | Receptor tyrosine-protein kinase erbB-2 | TK tyrosine-protein kinase EGFR subfamily | C00002526 | 5 / 9 |
| P20309 | Muscarinic acetylcholine receptor M3 | Acetylcholine receptor | C00002526 | 1 / 0 |
| P21452 | Substance-K receptor | Neurokinin receptor | C00002526 | 0 / 0 |
| P12931 | Proto-oncogene tyrosine-protein kinase Src | Src | C00002526 | 0 / 0 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00002526 | 0 / 0 |
| P51681 | C-C chemokine receptor type 5 | CC chemokine receptor | C00002526 | 3 / 0 |
| P50406 | 5-hydroxytryptamine receptor 6 | Serotonin receptor | C00002526 | 0 / 0 |
| P04745 | Alpha-amylase 1 | Enzyme | C00002526 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00002526 | 0 / 0 |
| Q13133 | Oxysterols receptor LXR-alpha | NR1H3 | C00002526 | 0 / 0 |
| P41597 | C-C chemokine receptor type 2 | CC chemokine receptor | C00002526 | 1 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00002526 | 0 / 0 |
| P08575 | Receptor-type tyrosine-protein phosphatase C | Enzyme | C00002526 | 2 / 1 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00002526 | 0 / 0 |
| P55055 | Oxysterols receptor LXR-beta | NR1H3 | C00002526 | 0 / 0 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00002526 | 2 / 0 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00002526 | 1 / 1 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002526 | 2 / 2 |
| O15118 | Niemann-Pick C1 protein | Unclassified protein | C00002526 | 1 / 1 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00002526 | 5 / 2 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00002526 | 0 / 0 |
| O76074 | cGMP-specific 3',5'-cyclic phosphodiesterase | PDE_5A | C00002526 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00002526 | 1 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00002526 | 1 / 0 |
| P22303 | Acetylcholinesterase | Hydrolase | C00002526 | 1 / 0 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | C00002526 | 0 / 0 |
| P28335 | 5-hydroxytryptamine receptor 2C | Serotonin receptor | C00002526 | 0 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00002526 | 0 / 0 |
| P08173 | Muscarinic acetylcholine receptor M4 | Acetylcholine receptor | C00002526 | 0 / 0 |
| P25103 | Substance-P receptor | Neurokinin receptor | C00002526 | 0 / 0 |
| P25105 | Platelet-activating factor receptor | PAF receptor | C00002526 | 0 / 0 |
| P33032 | Melanocortin receptor 5 | Melanocortin receptor | C00002526 | 0 / 0 |
| O95718 | Steroid hormone receptor ERR2 | NR3B2 | C00002526 | 1 / 1 |
| Q02880 | DNA topoisomerase 2-beta | Isomerase | C00002526 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00002526 | 0 / 0 |
| Q8WUI4 | Histone deacetylase 7 | Hydrolase | C00002526 | 0 / 0 |
| P31749 | RAC-alpha serine/threonine-protein kinase | Akt | C00002526 | 4 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00002526 | 1 / 1 |
| P37059 | Estradiol 17-beta-dehydrogenase 2 | Enzyme | C00002526 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002526 | 0 / 1 |
| P05181 | Cytochrome P450 2E1 | Cytochrome P450 2E1 | C00002526 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00002526 | 4 / 3 |
| Q99549 | M-phase phosphoprotein 8 | Unclassified protein | C00002526 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00002526 | 0 / 0 |
| O43451 | Maltase-glucoamylase, intestinal | Hydrolase | C00002526 | 0 / 0 |
| P10275 | Androgen receptor | NR3C4 | C00002526 | 3 / 4 |
| P23975 | Sodium-dependent noradrenaline transporter | Norepinephrine | C00002526 | 1 / 1 |
| P25100 | Alpha-1D adrenergic receptor | Adrenergic receptor | C00002526 | 0 / 0 |
| P30542 | Adenosine receptor A1 | Adenosine receptor | C00002526 | 0 / 0 |
| P18089 | Alpha-2B adrenergic receptor | Adrenergic receptor | C00002526 | 0 / 0 |
| P24557 | Thromboxane-A synthase | Cytochrome P450 5A1 | C00002526 | 1 / 1 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00002526 | 0 / 1 |
| P35869 | Aryl hydrocarbon receptor | Transcription Factor | C00002526 | 0 / 0 |
| P25025 | C-X-C chemokine receptor type 2 | CXC chemokine receptor | C00002526 | 0 / 0 |
| P43405 | Tyrosine-protein kinase SYK | Syk | C00002526 | 0 / 0 |
| Q06278 | Aldehyde oxidase | Enzyme | C00002526 | 0 / 1 |
| P40225 | Thrombopoietin | Unclassified protein | C00002526 | 1 / 1 |
| P56937 | 3-keto-steroid reductase | Enzyme | C00002526 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002526 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00002526 | 1 / 1 |
| O00255 | Menin | Unclassified protein | C00002526 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00002526 | 1 / 2 |
| P27338 | Amine oxidase [flavin-containing] B | Oxidoreductase | C00002526 | 0 / 0 |
| P42684 | Abelson tyrosine-protein kinase 2 | Abl | C00002526 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002526 | 0 / 0 |
1126 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 18 | ABAT, GABA-AT, GABAT, NPD009 | 4-aminobutyrate aminotransferase (EC:2.6.1.19 2.6.1.22) |
C00002526
|
| 19 | ABCA1, ABC-1, ABC1, CERP, HDLDT1, TGD | ATP-binding cassette, sub-family A (ABC1), member 1 |
C00002526
|
| 5243 | ABCB1, ABC20, CD243, CLCS, GP170, MDR1, P-GP, PGY1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 (EC:3.6.3.44) |
C00002526
|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00002526
|
| 85320 | ABCC11, EWWD, MRP8, WW | ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
C00002526
|
| 150000 | ABCC13, C21orf73, PRED6 | ATP-binding cassette, sub-family C (CFTR/MRP), member 13, pseudogene |
C00002526
|
| 1244 | ABCC2, ABC30, CMOAT, DJS, MRP2, cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
C00002526
|
| 8714 | ABCC3, ABC31, EST90757, MLP2, MOAT-D, MRP3, cMOAT2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
C00002526
|
| 83451 | ABHD11, WBSCR21 | abhydrolase domain containing 11 |
C00002526
|
| 10006 | ABI1, ABI-1, ABLBP4, E3B1, NAP1BP, SSH3BP, SSH3BP1 | abl-interactor 1 |
C00002526
|
| 1238 | ACKR2, CCBP2, CCR10, CCR9, CMKBR9, D6, hD6 | atypical chemokine receptor 2 |
C00002526
|
| 47 | ACLY, ACL, ATPCL, CLATP | ATP citrate lyase (EC:2.3.3.8) |
C00002526
|
| 51 | ACOX1, ACOX, PALMCOX, SCOX | acyl-CoA oxidase 1, palmitoyl (EC:1.3.3.6) |
C00002526
|
| 55 | ACPP, 5'-NT, ACP-3, ACP3, PAP | acid phosphatase, prostate (EC:3.1.3.2 3.1.3.5) |
C00002526
|
| 2181 | ACSL3, ACS3, FACL3, PRO2194 | acyl-CoA synthetase long-chain family member 3 (EC:6.2.1.3) |
C00002526
|
| 59 | ACTA2, AAT6, ACTSA, MYMY5 | actin, alpha 2, smooth muscle, aorta |
C00002526
|
| 345651 | ACTBL2, ACT | actin, beta-like 2 |
C00002526
|
| 95 | ACY1, ACY-1, ACY1D | aminoacylase 1 (EC:3.5.1.14) |
C00002526
|
| 8751 | ADAM15, MDC15 | ADAM metallopeptidase domain 15 |
C00002526
|
| 11173 | ADAMTS7, ADAM-TS_7, ADAM-TS7, ADAMTS-7 | ADAM metallopeptidase with thrombospondin type 1 motif, 7 (EC:3.4.24.-) |
C00002526
|
| 56999 | ADAMTS9 | ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
C00002526
|
| 119 | ADD2, ADDB | adducin 2 (beta) |
C00002526
|
| 10974 | ADIRF, AFRO, APM2, C10orf116, apM-2 | adipogenesis regulatory factor |
C00002526
|
| 152 | ADRA2C, ADRA2L2, ADRA2RL2, ADRARL2, ALPHA2CAR | adrenoceptor alpha 2C |
C00002526
|
| 158 | ADSL, AMPS, ASASE, ASL | adenylosuccinate lyase (EC:4.3.2.2) |
C00002526
|
| 10555 | AGPAT2, 1-AGPAT2, BSCL, BSCL1, LPAAB, LPAAT-beta | 1-acylglycerol-3-phosphate O-acyltransferase 2 (EC:2.3.1.51) |
C00002526
|
| 10551 | AGR2, AG2, GOB-4, HAG-2, PDIA17, XAG-2 | anterior gradient 2 |
C00002526
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002526
|
| 205 | AK4, AK_4, AK3, AK3L1, AK3L2 | adenylate kinase 4 (EC:2.7.4.10 2.7.4.6) |
C00002526
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00002526
|
| 208 | AKT2, HIHGHH, PKBB, PKBBETA, PRKBB, RAC-BETA | v-akt murine thymoma viral oncogene homolog 2 (EC:2.7.11.1) |
C00002526
|
| 220 | ALDH1A3, ALDH1A6, ALDH6, MCOP8, RALDH3 | aldehyde dehydrogenase 1 family, member A3 (EC:1.2.1.5) |
C00002526
|
| 224 | ALDH3A2, ALDH10, FALDH, SLS | aldehyde dehydrogenase 3 family, member A2 (EC:1.2.1.3) |
C00002526
|
| 229 | ALDOB, ALDB, ALDO2 | aldolase B, fructose-bisphosphate (EC:4.1.2.13) |
C00002526
|
| 248 | ALPI, IAP | alkaline phosphatase, intestinal (EC:3.1.3.1) |
C00002526
|
| 249 | ALPL, AP-TNAP, APTNAP, HOPS, TNAP, TNSALP | alkaline phosphatase, liver/bone/kidney (EC:3.1.3.1) |
C00002526
|
| 250 | ALPP, ALP, PALP, PLAP, PLAP-1 | alkaline phosphatase, placental (EC:3.1.3.1) |
C00002526
|
| 251 | ALPPL2, ALPG, ALPPL, GCAP | alkaline phosphatase, placental-like 2 (EC:3.1.3.1) |
C00002526
|
| 196394 | AMN1 | antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
C00002526
|
| 271 | AMPD2 | adenosine monophosphate deaminase 2 (EC:3.5.4.6) |
C00002526
|
| 64682 | ANAPC1, APC1, MCPR, TSG24 | anaphase promoting complex subunit 1 |
C00002526
|
| 284 | ANGPT1, AGP1, AGPT, ANG1 | angiopoietin 1 |
C00002526
|
| 286 | ANK1, ANK, SPH1, SPH2 | ankyrin 1, erythrocytic |
C00002526
|
| 54522 | ANKRD16 | ankyrin repeat domain 16 |
C00002526
|
| 91074 | ANKRD30A, NY-BR-1 | ankyrin repeat domain 30A |
C00002526
|
| 57730 | ANKRD36B, KIAA1641 | ankyrin repeat domain 36B |
C00002526
|
| 55107 | ANO1, DOG1, ORAOV2, TAOS2, TMEM16A | anoctamin 1, calcium activated chloride channel |
C00002526
|
| 8125 | ANP32A, C15orf1, HPPCn, I1PP2A, LANP, MAPM, PHAP1, PHAPI, PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
C00002526
|
| 301 | ANXA1, ANX1, LPC1 | annexin A1 |
C00002526
|
| 306 | ANXA3, ANX3 | annexin A3 (EC:3.1.4.43) |
C00002526
|
| 8905 | AP1S2, MGC:1902, MRX59, MRXS21, MRXSF, SIGMA1B | adaptor-related protein complex 1, sigma 2 subunit |
C00002526
|
| 317 | APAF1, APAF-1, CED4 | apoptotic peptidase activating factor 1 |
C00002526
|
| 334 | APLP2, APLP-2, APPH, APPL2, CDEBP | amyloid beta (A4) precursor-like protein 2 |
C00002526
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00002526
|
| 347 | APOD | apolipoprotein D |
C00002526
|
| 353 | APRT, AMP, APRTD | adenine phosphoribosyltransferase (EC:2.4.2.7) |
C00002526
|
| 358 | AQP1, AQP-CHIP, CHIP28, CO | aquaporin 1 |
C00002526
|
| 360 | AQP3, AQP-3, GIL | aquaporin 3 (Gill blood group) |
C00002526
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00002526
|
| 374 | AREG, AR, CRDGF, SDGF | amphiregulin |
C00002526
|
| 375 | ARF1 | ADP-ribosylation factor 1 |
C00002526
|
| 396 | ARHGDIA, GDIA1, NPHS8, RHOGDI, RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha |
C00002526
|
| 403 | ARL3, ARFL3 | ADP-ribosylation factor-like 3 |
C00002526
|
| 83787 | ARMC10, PNAS-112, PNAS112, SVH | armadillo repeat containing 10 |
C00002526
|
| 51566 | ARMCX3, ALEX3, dJ545K15.2 | armadillo repeat containing, X-linked 3 |
C00002526
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002526
|
| 414 | ARSD, ASD | arylsulfatase D (EC:3.1.6.1) |
C00002526
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00002526
|
| 440 | ASNS, TS11 | asparagine synthetase (glutamine-hydrolyzing) (EC:6.3.5.4) |
C00002526
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00002526
|
| 80150 | ASRGL1, ALP, ALP1, CRASH | asparaginase like 1 (EC:3.5.1.1 3.4.19.5) |
C00002526
|
| 445 | ASS1, ASS, CTLN1 | argininosuccinate synthase 1 (EC:6.3.4.5) |
C00002526
|
| 29028 | ATAD2, ANCCA, PRO2000 | ATPase family, AAA domain containing 2 (EC:3.6.1.3) |
C00002526
|
| 467 | ATF3 | activating transcription factor 3 |
C00002526
|
| 9474 | ATG5, APG5, APG5-LIKE, APG5L, ASP, hAPG5 | autophagy related 5 |
C00002526
|
| 472 | ATM, AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | ataxia telangiectasia mutated (EC:2.7.11.1) |
C00002526
|
| 1822 | ATN1, B37, D12S755E, DRPLA, HRS, NOD | atrophin 1 |
C00002526
|
| 481 | ATP1B1, ATP1B | ATPase, Na+/K+ transporting, beta 1 polypeptide |
C00002526
|
| 514 | ATP5E, ATPE, MC5DN3 | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit (EC:3.6.3.14) |
C00002526
|
| 23545 | ATP6V0A2, A2, ARCL, ARCL2A, ATP6A2, ATP6N1D, J6B7, RTF, STV1, TJ6, TJ6M, TJ6S, VPH1, WSS | ATPase, H+ transporting, lysosomal V0 subunit a2 (EC:3.6.3.6) |
C00002526
|
| 11273 | ATXN2L, A2D, A2LG, A2LP, A2RP | ataxin 2-like |
C00002526
|
| 6790 | AURKA, AIK, ARK1, AURA, AURORA2, BTAK, PPP1R47, STK15, STK6, STK7 | aurora kinase A (EC:2.7.11.1) |
C00002526
|
| 9212 | AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, PPP1R48, STK12, STK5, aurkb-sv1, aurkb-sv2 | aurora kinase B (EC:2.7.11.1) |
C00002526
|
| 2683 | B4GALT1, B4GAL-T1, CDG2D, GGTB2, GT1, GTB, beta4Gal-T1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (EC:2.4.1.22 2.4.1.90 2.4.1.38) |
C00002526
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00002526
|
| 9529 | BAG5, BAG-5 | BCL2-associated athanogene 5 |
C00002526
|
| 10409 | BASP1, CAP-23, CAP23, NAP-22, NAP22 | brain abundant, membrane attached signal protein 1 |
C00002526
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00002526
|
| 11177 | BAZ1A, ACF1, WALp1, WCRF180, hACF1 | bromodomain adjacent to zinc finger domain, 1A |
C00002526
|
| 11176 | BAZ2A, TIP5, WALp3 | bromodomain adjacent to zinc finger domain, 2A |
C00002526
|
| 590 | BCHE, CHE1, CHE2, E1 | butyrylcholinesterase (EC:3.1.1.8) |
C00002526
|
| 593 | BCKDHA, BCKDE1A, MSU, MSUD1, OVD1A | branched chain keto acid dehydrogenase E1, alpha polypeptide (EC:1.2.4.4) |
C00002526
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00002526
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00002526
|
| 632 | BGLAP, BGP, OC, OCN | bone gamma-carboxyglutamate (gla) protein |
C00002526
|
| 168620 | BHLHA15, BHLHB8, MIST1 | basic helix-loop-helix family, member a15 |
C00002526
|
| 8553 | BHLHE40, BHLHB2, DEC1, HLHB2, SHARP-2, STRA13, Stra14 | basic helix-loop-helix family, member e40 |
C00002526
|
| 636 | BICD1, BICD | bicaudal D homolog 1 (Drosophila) |
C00002526
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00002526
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00002526
|
| 90427 | BMF | Bcl2 modifying factor |
C00002526
|
| 662 | BNIP1, NIP1, SEC20, TRG-8 | BCL2/adenovirus E1B 19kDa interacting protein 1 |
C00002526
|
| 665 | BNIP3L, BNIP3a, NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like |
C00002526
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00002526
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00002526
|
| 55845 | BRK1, C3orf10, MDS027, hHBrk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit |
C00002526
|
| 428592 |
C00002526
|
||
| 103993 |
C00002526
|
||
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00002526
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00002526
|
| 10950 | BTG3, ANA, TOB5, TOB55, TOFA | BTG family, member 3 |
C00002526
|
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00002526
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00002526
|
| 56652 | C10orf2, ATXN8, IOSCA, MTDPS7, PEO, PEO1, PEOA3, SANDO, SCA8, TWINL | chromosome 10 open reading frame 2 (EC:3.6.4.12) |
C00002526
|
| 219833 | C11orf45 | chromosome 11 open reading frame 45 |
C00002526
|
| 84419 | C15orf48, FOAP-11, NMES1 | chromosome 15 open reading frame 48 |
C00002526
|
| 79415 | C17orf62 | chromosome 17 open reading frame 62 |
C00002526
|
| 388722 | C1orf53 | chromosome 1 open reading frame 53 |
C00002526
|
| 79680 | C22orf29, BOP | chromosome 22 open reading frame 29 |
C00002526
|
| 79919 | C2orf54 | chromosome 2 open reading frame 54 |
C00002526
|
| 718 | C3, AHUS5, ARMD9, ASP, C3a, C3b, CPAMD1 | complement component 3 (EC:3.4.21.43) |
C00002526
|
| 100495920 |
C00002526
|
||
| 50854 | C6orf48, D6S57, G8 | chromosome 6 open reading frame 48 |
C00002526
|
| 771 | CA12, CAXII, HsT18816 | carbonic anhydrase XII (EC:4.2.1.1) |
C00002526
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00002526
|
| 91768 | CABLES1, CABL1, CABLES, HsT2563, IK3-1 | Cdk5 and Abl enzyme substrate 1 |
C00002526
|
| 8911 | CACNA1I, Cav3.3, ca(v)3.3 | calcium channel, voltage-dependent, T type, alpha 1I subunit |
C00002526
|
| 55450 | CAMK2N1, PRO1489, RP11-401M16.1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
C00002526
|
| 10645 | CAMKK2, CAMKK, CAMKKB | calcium/calmodulin-dependent protein kinase kinase 2, beta (EC:2.7.11.17) |
C00002526
|
| 821 | CANX, CNX, IP90, P90 | calnexin |
C00002526
|
| 823 | CAPN1, CANP, CANP1, CANPL1, muCANP, muCL | calpain 1, (mu/I) large subunit (EC:3.4.22.52) |
C00002526
|
| 6650 | CAPN15, SOLH | calpain 15 |
C00002526
|
| 10753 | CAPN9, GC36, nCL-4 | calpain 9 (EC:3.4.22.-) |
C00002526
|
| 828 | CAPS, CAPS1 | calcyphosine |
C00002526
|
| 29775 | CARD10, BIMP1, CARMA3 | caspase recruitment domain family, member 10 |
C00002526
|
| 833 | CARS, CARS1, CYSRS, MGC:11246 | cysteinyl-tRNA synthetase (EC:6.1.1.16) |
C00002526
|
| 843 | CASP10, ALPS2, FLICE2, MCH4 | caspase 10, apoptosis-related cysteine peptidase (EC:3.4.22.63) |
C00002526
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00002526
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00002526
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00002526
|
| 9994 | CASP8AP2, CED-4, FLASH, RIP25 | caspase 8 associated protein 2 |
C00002526
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00002526
|
| 863 | CBFA2T3, ETO2, MTG16, MTGR2, ZMYND4 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
C00002526
|
| 873 | CBR1, CBR, SDR21C1, hCBR1 | carbonyl reductase 1 (EC:1.1.1.189 1.1.1.197 1.1.1.184) |
C00002526
|
| 23468 | CBX5, HP1, HP1A | chromobox homolog 5 |
C00002526
|
| 23466 | CBX6 | chromobox homolog 6 |
C00002526
|
| 55246 | CCDC25 | coiled-coil domain containing 25 |
C00002526
|
| 26112 | CCDC69 | coiled-coil domain containing 69 |
C00002526
|
| 79080 | CCDC86 | coiled-coil domain containing 86 |
C00002526
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002526
|
| 6351 | CCL4, ACT2, AT744.1, G-26, HC21, LAG-1, LAG1, MIP-1-beta, MIP1B, MIP1B1, SCYA2, SCYA4 | chemokine (C-C motif) ligand 4 |
C00002526
|
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00002526
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00002526
|
| 9133 | CCNB2, HsT17299 | cyclin B2 |
C00002526
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002526
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002526
|
| 960 | CD44, CDW44, CSPG8, ECMR-III, HCELL, HUTCH-I, IN, LHR, MC56, MDU2, MDU3, MIC4, Pgp1 | CD44 molecule (Indian blood group) |
C00002526
|
| 965 | CD58, LFA-3, LFA3, ag3 | CD58 molecule |
C00002526
|
| 3732 | CD82, 4F9, C33, GR15, IA4, KAI1, R2, SAR2, ST6, TSPAN27 | CD82 molecule |
C00002526
|
| 928 | CD9, BTCC-1, DRAP-27, MIC3, MRP-1, TSPAN-29, TSPAN29 | CD9 molecule |
C00002526
|
| 976 | CD97, TM7LN1 | CD97 molecule |
C00002526
|
| 991 | CDC20, CDC20A, bA276H19.3, p55CDC | cell division cycle 20 |
C00002526
|
| 994 | CDC25B | cell division cycle 25B (EC:3.1.3.48) |
C00002526
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00002526
|
| 11135 | CDC42EP1, BORG5, CEP1, MSE55 | CDC42 effector protein (Rho GTPase binding) 1 |
C00002526
|
| 8318 | CDC45, CDC45L, CDC45L2, PORC-PI-1 | cell division cycle 45 |
C00002526
|
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00002526
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00002526
|
| 1000 | CDH2, CD325, CDHN, CDw325, NCAD | cadherin 2, type 1, N-cadherin (neuronal) |
C00002526
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00002526
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002526
|
| 10263 | CDK2AP2, DOC-1R, p14 | cyclin-dependent kinase 2 associated protein 2 |
C00002526
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00002526
|
| 1020 | CDK5, PSSALRE | cyclin-dependent kinase 5 (EC:2.7.11.22) |
C00002526
|
| 1024 | CDK8, K35 | cyclin-dependent kinase 8 (EC:2.7.11.23 2.7.11.22) |
C00002526
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002526
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00002526
|
| 1028 | CDKN1C, BWCR, BWS, KIP2, WBS, p57, p57Kip2 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
C00002526
|
| 1031 | CDKN2C, INK4C, p18, p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
C00002526
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00002526
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00002526
|
| 1051 | CEBPB, C/EBP-beta, CRP2, IL6DBP, LAP, LIP, NF-IL6, TCF5 | CCAAT/enhancer binding protein (C/EBP), beta |
C00002526
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00002526
|
| 1952 | CELSR2, CDHF10, EGFL2, Flamingo1, MEGF3 | cadherin, EGF LAG seven-pass G-type receptor 2 |
C00002526
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00002526
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00002526
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00002526
|
| 55835 | CENPJ, BM032, CENP-J, CPAP, LAP, LIP1, MCPH6, SASS4, SCKL4, Sas-4 | centromere protein J |
C00002526
|
| 55839 | CENPN, BM039, C16orf60, CENP-N, ICEN32 | centromere protein N |
C00002526
|
| 1080 | CFTR, ABC35, ABCC7, CF, CFTR/MRP, MRP7, TNR-CFTR, dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) (EC:3.6.3.49) |
C00002526
|
| 494143 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) |
C00002526
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00002526
|
| 118487 | CHCHD1, C10orf34, C2360, MRP-S37 | coiled-coil-helix-coiled-coil-helix domain containing 1 |
C00002526
|
| 1111 | CHEK1, CHK1 | checkpoint kinase 1 (EC:2.7.11.1) |
C00002526
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00002526
|
| 25978 | CHMP2B, ALS17, CHMP2.5, DMT1, VPS2-2, VPS2B | charged multivesicular body protein 2B |
C00002526
|
| 1124 | CHN2, ARHGAP3, BCH, CHN2-3, RHOGAP3 | chimerin 2 |
C00002526
|
| 22856 | CHSY1, CHSY, CSS1, ChSy-1, TPBS | chondroitin sulfate synthase 1 (EC:2.4.1.175 2.4.1.226) |
C00002526
|
| 8483 | CILP, CILP-1, HsT18872 | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
C00002526
|
| 1154 | CISH, BACTS2, CIS, CIS-1, G18, SOCS | cytokine inducible SH2-containing protein |
C00002526
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00002526
|
| 10370 | CITED2, ASD8, MRG-1, MRG1, P35SRJ, VSD2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
C00002526
|
| 1152 | CKB, B-CK, CKBB | creatine kinase, brain (EC:2.7.3.2) |
C00002526
|
| 1163 | CKS1B, CKS1, PNAS-16, PNAS-18, ckshs1 | CDC28 protein kinase regulatory subunit 1B |
C00002526
|
| 9076 | CLDN1, CLD1, ILVASC, SEMP1 | claudin 1 |
C00002526
|
| 10686 | CLDN16, HOMG3, PCLN1 | claudin 16 |
C00002526
|
| 1364 | CLDN4, CPE-R, CPER, CPETR, CPETR1, WBSCR8, hCPE-R | claudin 4 |
C00002526
|
| 54102 | CLIC6, CLIC1L | chloride intracellular channel 6 |
C00002526
|
| 1209 | CLPTM1 | cleft lip and palate associated transmembrane protein 1 |
C00002526
|
| 23277 | CLUH, CLU1, KIAA0664 | clustered mitochondria (cluA/CLU1) homolog |
C00002526
|
| 112616 | CMTM7, CKLFSF7 | CKLF-like MARVEL transmembrane domain containing 7 |
C00002526
|
| 84518 | CNFN, PLAC8L2 | cornifelin |
C00002526
|
| 1264 | CNN1, SMCC, Sm-Calp | calponin 1, basic, smooth muscle |
C00002526
|
| 1265 | CNN2 | calponin 2 |
C00002526
|
| 10330 | CNPY2, HP10390, MSAP, TMEM4, ZSIG9 | canopy FGF signaling regulator 2 |
C00002526
|
| 1303 | COL12A1, BA209D8.1, COL12A1L, DJ234P15.1 | collagen, type XII, alpha 1 |
C00002526
|
| 80781 | COL18A1, KNO, KNO1, KS | collagen, type XVIII, alpha 1 |
C00002526
|
| 1291 | COL6A1, OPLL | collagen, type VI, alpha 1 |
C00002526
|
| 1312 | COMT | catechol-O-methyltransferase (EC:2.1.1.6) |
C00002526
|
| 118881 | COMTD1 | catechol-O-methyltransferase domain containing 1 |
C00002526
|
| 23603 | CORO1C, HCRNN4 | coronin, actin binding protein, 1C |
C00002526
|
| 23406 | COTL1, CLP | coactosin-like 1 (Dictyostelium) |
C00002526
|
| 4512 | COX1, COI, MTCO1, MT-CO1 | cytochrome c oxidase subunit I |
C00002526
|
| 9167 | COX7A2L, COX7AR, COX7RP, EB1, SIG81 | cytochrome c oxidase subunit VIIa polypeptide 2 like |
C00002526
|
| 1357 | CPA1, CPA | carboxypeptidase A1 (pancreatic) (EC:3.4.17.1) |
C00002526
|
| 1374 | CPT1A, CPT1, CPT1-L, L-CPT1 | carnitine palmitoyltransferase 1A (liver) (EC:2.3.1.21) |
C00002526
|
| 1396 | CRIP1, CRHP, CRIP, CRP-1, CRP1 | cysteine-rich protein 1 (intestinal) |
C00002526
|
| 83690 | CRISPLD1, CRISP-10, CRISP10, LCRISP1 | cysteine-rich secretory protein LCCL domain containing 1 |
C00002526
|
| 1401 | CRP, PTX1 | C-reactive protein, pentraxin-related |
C00002526
|
| 445748 |
C00002526
|
||
| 1465 | CSRP1, CRP, CRP1, CSRP, CYRP, D1S181E | cysteine and glycine-rich protein 1 |
C00002526
|
| 57325 | CSRP2BP, ATAC2, CRP2BP, KAT14, PRO1194, dJ717M23.1 | CSRP2 binding protein |
C00002526
|
| 1475 | CSTA, AREI, STF1, STFA | cystatin A (stefin A) |
C00002526
|
| 1488 | CTBP2 | C-terminal binding protein 2 |
C00002526
|
| 1490 | CTGF, CCN2, HCS24, IGFBP8, NOV2 | connective tissue growth factor |
C00002526
|
| 1501 | CTNND2, GT24, NPRAP | catenin (cadherin-associated protein), delta 2 |
C00002526
|
| 1503 | CTPS1, CTPS | CTP synthase 1 (EC:6.3.4.2) |
C00002526
|
| 1075 | CTSC, CPPI, DPP-I, DPP1, DPPI, HMS, JP, JPD, PALS, PDON1, PLS | cathepsin C (EC:3.4.14.1) |
C00002526
|
| 1509 | CTSD, CLN10, CPSD | cathepsin D (EC:3.4.23.5) |
C00002526
|
| 8453 | CUL2 | cullin 2 |
C00002526
|
| 51596 | CUTA, ACHAP, C6orf82 | cutA divalent cation tolerance homolog (E. coli) |
C00002526
|
| 1523 | CUX1, CASP, CDP, CDP/Cut, CDP1, COY1, CUTL1, CUX, Clox, Cux/CDP, GOLIM6, Nbla10317, p100, p110, p200, p75 | cut-like homeobox 1 |
C00002526
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00002526
|
| 9547 | CXCL14, BMAC, BRAK, KEC, KS1, MIP-2g, MIP2G, NJAC, SCYB14 | chemokine (C-X-C motif) ligand 14 |
C00002526
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00002526
|
| 1528 | CYB5A, CYB5, MCB5 | cytochrome b5 type A (microsomal) |
C00002526
|
| 1535 | CYBA, p22-PHOX | cytochrome b-245, alpha polypeptide |
C00002526
|
| 54205 | CYCS, CYC, HCS, THC4 | cytochrome c, somatic |
C00002526
|
| 114757 | CYGB, HGB, STAP | cytoglobin |
C00002526
|
| 1540 | CYLD, BRSS, CDMT, CYLD1, CYLDI, EAC, MFT, MFT1, SBS, TEM, USPL2 | cylindromatosis (turban tumor syndrome) (EC:3.4.19.12) |
C00002526
|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002526
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002526
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00002526
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00002526
|
| 1591 | CYP24A1, CP24, CYP24, HCAI, P450-CC24 | cytochrome P450, family 24, subfamily A, polypeptide 1 (EC:1.14.13.126) |
C00002526
|
| 1594 | CYP27B1, CP2B, CYP1, CYP1alpha, CYP27B, P450c1, PDDR, VDD1, VDDR, VDDRI, VDR | cytochrome P450, family 27, subfamily B, polypeptide 1 (EC:1.14.13.13) |
C00002526
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00002526
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00002526
|
| 10858 | CYP46A1, CP46, CYP46 | cytochrome P450, family 46, subfamily A, polypeptide 1 (EC:1.14.13.98) |
C00002526
|
| 1580 | CYP4B1, CYPIVB1, P-450HP | cytochrome P450, family 4, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00002526
|
| 8529 | CYP4F2, CPF2 | cytochrome P450, family 4, subfamily F, polypeptide 2 (EC:1.14.13.30) |
C00002526
|
| 3491 | CYR61, CCN1, GIG1, IGFBP10 | cysteine-rich, angiogenic inducer, 61 |
C00002526
|
| 9267 | CYTH1, B2-1, CYTOHESIN-1, D17S811E, PSCD1, SEC7 | cytohesin 1 |
C00002526
|
| 9265 | CYTH3, ARNO3, GRP1, PSCD3 | cytohesin 3 |
C00002526
|
| 1603 | DAD1, OST2 | defender against cell death 1 (EC:2.4.99.18) |
C00002526
|
| 55861 | DBNDD2, C20orf35, CK1BP, HSMNP1 | dysbindin (dystrobrevin binding protein 1) domain containing 2 |
C00002526
|
| 1638 | DCT, TRP-2, TYRP2 | dopachrome tautomerase (EC:5.3.3.12) |
C00002526
|
| 51181 | DCXR, DCR, HCR2, HCRII, KIDCR, P34H, SDR20C1, XR | dicarbonyl/L-xylulose reductase (EC:1.1.1.10) |
C00002526
|
| 1644 | DDC, AADC | dopa decarboxylase (aromatic L-amino acid decarboxylase) (EC:4.1.1.28) |
C00002526
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00002526
|
| 65992 | DDRGK1, C20orf116, UFBP1, dJ1187M17.3 | DDRGK domain containing 1 |
C00002526
|
| 123099 | DEGS2, C14orf66, DES2, FADS8 | delta(4)-desaturase, sphingolipid 2 |
C00002526
|
| 55789 | DEPDC1B, BRCC3, XTP1 | DEP domain containing 1B |
C00002526
|
| 420357 |
C00002526
|
||
| 1677 | DFFB, CAD, CPAN, DFF-40, DFF2, DFF40 | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
C00002526
|
| 84649 | DGAT2, ARAT, GS1999FULL | diacylglycerol O-acyltransferase 2 (EC:2.3.1.20 2.3.1.76) |
C00002526
|
| 8526 | DGKE, DAGK5, DAGK6, DGK, NPHS7 | diacylglycerol kinase, epsilon 64kDa (EC:2.7.1.107) |
C00002526
|
| 1718 | DHCR24, DCE, Nbla03646, SELADIN1, seladin-1 | 24-dehydrocholesterol reductase (EC:1.3.1.72) |
C00002526
|
| 200895 | DHFRL1, DHFRP4 | dihydrofolate reductase-like 1 (EC:1.5.1.3) |
C00002526
|
| 308585 |
C00002526
|
||
| 10202 | DHRS2, HEP27, SDR25C1 | dehydrogenase/reductase (SDR family) member 2 |
C00002526
|
| 9249 | DHRS3, DD83.1, RDH17, Rsdr1, SDR1, SDR16C1, retSDR1 | dehydrogenase/reductase (SDR family) member 3 (EC:1.1.1.300) |
C00002526
|
| 22943 | DKK1, DKK-1, SK | dickkopf WNT signaling pathway inhibitor 1 |
C00002526
|
| 9231 | DLG5, LP-DLG, P-DLG5, PDLG | discs, large homolog 5 (Drosophila) |
C00002526
|
| 9787 | DLGAP5, DLG7, HURP | discs, large (Drosophila) homolog-associated protein 5 |
C00002526
|
| 1756 | DMD, BMD, CMD3B, DXS142, DXS164, DXS206, DXS230, DXS239, DXS268, DXS269, DXS270, DXS272 | dystrophin |
C00002526
|
| 4189 | DNAJB9, ERdj4, MDG-1, MDG1, MST049, MSTP049 | DnaJ (Hsp40) homolog, subfamily B, member 9 |
C00002526
|
| 55192 | DNAJC17 | DnaJ (Hsp40) homolog, subfamily C, member 17 |
C00002526
|
| 134218 | DNAJC21, DNAJA5, GS3, JJJ1 | DnaJ (Hsp40) homolog, subfamily C, member 21 |
C00002526
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00002526
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00002526
|
| 23549 | DNPEP, ASPEP, DAP | aspartyl aminopeptidase (EC:3.4.11.21) |
C00002526
|
| 1795 | DOCK3, MOCA, PBP | dedicator of cytokinesis 3 |
C00002526
|
| 83475 | DOHH, HLRC1, hDOHH | deoxyhypusine hydroxylase/monooxygenase (EC:1.14.99.29) |
C00002526
|
| 55715 | DOK4 | docking protein 4 |
C00002526
|
| 285489 | DOK7, C4orf25, CMS1B | docking protein 7 |
C00002526
|
| 1819 | DRG2 | developmentally regulated GTP binding protein 2 |
C00002526
|
| 1830 | DSG3, CDHF6, PVA | desmoglein 3 |
C00002526
|
| 51514 | DTL, CDT2, DCAF2, L2DTL, RAMP | denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
C00002526
|
| 23220 | DTX4, RNF155 | deltex homolog 4 (Drosophila) |
C00002526
|
| 1841 | DTYMK, CDC8, PP3731, TMPK, TYMK | deoxythymidylate kinase (thymidylate kinase) (EC:2.7.4.9) |
C00002526
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00002526
|
| 1844 | DUSP2, PAC-1, PAC1 | dual specificity phosphatase 2 (EC:3.1.3.16 3.1.3.48) |
C00002526
|
| 1846 | DUSP4, HVH2, MKP-2, MKP2, TYP | dual specificity phosphatase 4 (EC:3.1.3.16 3.1.3.48) |
C00002526
|
| 1847 | DUSP5, DUSP, HVH3 | dual specificity phosphatase 5 (EC:3.1.3.16 3.1.3.48) |
C00002526
|
| 1848 | DUSP6, HH19, MKP3, PYST1 | dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48) |
C00002526
|
| 1855 | DVL1, DVL, DVL1L1, DVL1P1 | dishevelled segment polarity protein 1 |
C00002526
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00002526
|
| 9166 | EBAG9, EB9, PDAF | estrogen receptor binding site associated, antigen, 9 |
C00002526
|
| 10682 | EBP, CDPX2, CHO2, CPX, CPXD | emopamil binding protein (sterol isomerase) (EC:5.3.3.5) |
C00002526
|
| 1889 | ECE1, ECE | endothelin converting enzyme 1 (EC:3.4.24.71) |
C00002526
|
| 1891 | ECH1, HPXEL | enoyl CoA hydratase 1, peroxisomal |
C00002526
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00002526
|
| 23644 | EDC4, GE1, Ge-1, HEDL5, HEDLS, RCD-8, RCD8 | enhancer of mRNA decapping 4 |
C00002526
|
| 1906 | EDN1, ET1, HDLCQ7, PPET1 | endothelin 1 |
C00002526
|
| 1907 | EDN2, ET2, PPET2 | endothelin 2 |
C00002526
|
| 1938 | EEF2, EEF-2, EF-2, EF2, SCA26 | eukaryotic translation elongation factor 2 |
C00002526
|
| 80303 | EFHD1, MST133, MSTP133, SWS2 | EF-hand domain family, member D1 |
C00002526
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00002526
|
| 1950 | EGF, HOMG4, URG | epidermal growth factor |
C00002526
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00002526
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00002526
|
| 1960 | EGR3, EGR-3, PILOT | early growth response 3 |
C00002526
|
| 1981 | EIF4G1, EIF-4G1, EIF4F, EIF4G, EIF4GI, P220, PARK18 | eukaryotic translation initiation factor 4 gamma, 1 |
C00002526
|
| 1984 | EIF5A, EIF-5A, EIF5A1, eIF5AI | eukaryotic translation initiation factor 5A |
C00002526
|
| 64834 | ELOVL1, Ssc1 | ELOVL fatty acid elongase 1 (EC:2.3.1.199) |
C00002526
|
| 83401 | ELOVL3, CIG-30, CIG30 | ELOVL fatty acid elongase 3 (EC:2.3.1.199) |
C00002526
|
| 60481 | ELOVL5, HELO1, dJ483K16.1 | ELOVL fatty acid elongase 5 (EC:2.3.1.199) |
C00002526
|
| 2013 | EMP2, XMP | epithelial membrane protein 2 |
C00002526
|
| 2026 | ENO2, NSE | enolase 2 (gamma, neuronal) (EC:4.2.1.11) |
C00002526
|
| 2027 | ENO3, GSD13, MSE | enolase 3 (beta, muscle) (EC:4.2.1.11) |
C00002526
|
| 5168 | ENPP2, ATX, ATX-X, AUTOTAXIN, LysoPLD, NPP2, PD-IALPHA, PDNP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 (EC:3.1.4.39) |
C00002526
|
| 2036 | EPB41L1, 4.1N, MRD11 | erythrocyte membrane protein band 4.1-like 1 |
C00002526
|
| 2037 | EPB41L2, 4.1-G, 4.1G | erythrocyte membrane protein band 4.1-like 2 |
C00002526
|
| 2041 | EPHA1, EPH, EPHT, EPHT1 | EPH receptor A1 (EC:2.7.10.1) |
C00002526
|
| 2050 | EPHB4, HTK, MYK1, TYRO11 | EPH receptor B4 (EC:2.7.10.1) |
C00002526
|
| 2052 | EPHX1, EPHX, EPOX, HYL1, MEH | epoxide hydrolase 1, microsomal (xenobiotic) (EC:3.3.2.9) |
C00002526
|
| 2053 | EPHX2, CEH, SEH | epoxide hydrolase 2, cytoplasmic (EC:3.3.2.10 3.1.3.76) |
C00002526
|
| 2057 | EPOR, EPO-R | erythropoietin receptor |
C00002526
|
| 2068 | ERCC2, COFS2, EM9, TTD, XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 (EC:3.6.4.12) |
C00002526
|
| 2071 | ERCC3, BTF2, GTF2H, RAD25, TFIIH, XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (EC:3.6.4.12) |
C00002526
|
| 1161 | ERCC8, CKN1, CSA, UVSS2 | excision repair cross-complementing rodent repair deficiency, complementation group 8 |
C00002526
|
| 11160 | ERLIN2, C8orf2, Erlin-2, NET32, SPFH2, SPG18 | ER lipid raft associated 2 |
C00002526
|
| 83715 | ESPN, DFNB36 | espin |
C00002526
|
| 284729 | ESPNP, RP1-163M9.1, dJ1182A14.1 | espin pseudogene |
C00002526
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00002526
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00002526
|
| 2101 | ESRRA, ERR1, ERRa, ERRalpha, ESRL1, NR3B1 | estrogen-related receptor alpha |
C00002526
|
| 2103 | ESRRB, DFNB35, ERR2, ERRb, ESRL2, NR3B2 | estrogen-related receptor beta |
C00002526
|
| 2104 | ESRRG, ERR3, ERRgamma, NR3B3 | estrogen-related receptor gamma |
C00002526
|
| 55224 | ETNK2, EKI2, HMFT1716 | ethanolamine kinase 2 (EC:2.7.1.82) |
C00002526
|
| 2114 | ETS2, ETS2IT1 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
C00002526
|
| 2137 | EXTL3, BOTV, EXTL1L, EXTR1, REGR, RPR | exostosin-like glycosyltransferase 3 (EC:2.4.1.223) |
C00002526
|
| 9415 | FADS2, D6D, DES6, FADSD6, LLCDL2, SLL0262, TU13 | fatty acid desaturase 2 (EC:1.14.19.-) |
C00002526
|
| 399665 | FAM102A, C9orf132, EEIG1, SYM-3A, bA203J24.7 | family with sequence similarity 102, member A |
C00002526
|
| 11170 | FAM107A, DRR1, TU3A | family with sequence similarity 107, member A |
C00002526
|
| 116224 | FAM122A, C9orf42 | family with sequence similarity 122A |
C00002526
|
| 116496 | FAM129A, C1orf24, NIBAN | family with sequence similarity 129, member A |
C00002526
|
| 23201 | FAM168A, KIAA0280, TCRP1 | family with sequence similarity 168, member A |
C00002526
|
| 84331 | FAM195A, C16orf14, c349E10.1 | family with sequence similarity 195, member A |
C00002526
|
| 131583 | FAM43A | family with sequence similarity 43, member A |
C00002526
|
| 81610 | FAM83D, C20orf129, CHICA, dJ616B8.3 | family with sequence similarity 83, member D |
C00002526
|
| 157638 | FAM84B, BCMP101, NSE2 | family with sequence similarity 84, member B |
C00002526
|
| 2176 | FANCC, FA3, FAC, FACC | Fanconi anemia, complementation group C |
C00002526
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00002526
|
| 10160 | FARP1, CDEP, PLEKHC2, PPP1R75 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
C00002526
|
| 2192 | FBLN1, FBLN, FIBL1 | fibulin 1 |
C00002526
|
| 2200 | FBN1, ACMICD, ECTOL1, FBN, GPHYSD2, MASS, MFS1, OCTD, SGS, SSKS, WMS, WMS2 | fibrillin 1 |
C00002526
|
| 2201 | FBN2, CCA, DA9 | fibrillin 2 |
C00002526
|
| 114907 | FBXO32, Fbx32, MAFbx | F-box protein 32 |
C00002526
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00002526
|
| 2237 | FEN1, FEN-1, MF1, RAD2 | flap structure-specific endonuclease 1 |
C00002526
|
| 2246 | FGF1, AFGF, ECGF, ECGF-beta, ECGFA, ECGFB, FGF-1, FGF-alpha, FGFA, GLIO703, HBGF-1, HBGF1 | fibroblast growth factor 1 (acidic) |
C00002526
|
| 2263 | FGFR2, BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 | fibroblast growth factor receptor 2 (EC:2.7.10.1) |
C00002526
|
| 2274 | FHL2, AAG11, DRAL, FHL-2, SLIM-3, SLIM3 | four and a half LIM domains 2 |
C00002526
|
| 2288 | FKBP4, FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | FK506 binding protein 4, 59kDa (EC:5.2.1.8) |
C00002526
|
| 2316 | FLNA, ABP-280, ABPX, CSBS, CVD1, FLN, FLN-A, FLN1, FMD, MNS, NHBP, OPD, OPD1, OPD2, XLVD, XMVD | filamin A, alpha |
C00002526
|
| 56776 | FMN2 | formin 2 |
C00002526
|
| 252995 | FNDC5, FRCP2, irisin | fibronectin type III domain containing 5 |
C00002526
|
| 2348 | FOLR1, FBP, FOLR | folate receptor 1 (adult) |
C00002526
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00002526
|
| 2297 | FOXD1, FKHL8, FREAC4 | forkhead box D1 |
C00002526
|
| 2302 | FOXJ1, FKHL13, HFH-4, HFH4 | forkhead box J1 |
C00002526
|
| 2309 | FOXO3, AF6q21, FKHRL1, FKHRL1P2, FOXO2, FOXO3A | forkhead box O3 |
C00002526
|
| 2525 | FUT3, CD174, FT3B, FucT-III, LE, Les | fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (EC:2.4.1.65) |
C00002526
|
| 2527 | FUT5, FUC-TV | fucosyltransferase 5 (alpha (1,3) fucosyltransferase) (EC:2.4.1.65) |
C00002526
|
| 7976 | FZD3, Fz-3 | frizzled family receptor 3 |
C00002526
|
| 7855 | FZD5, C2orf31, HFZ5 | frizzled family receptor 5 |
C00002526
|
| 50486 | G0S2, RP1-28O10.2 | G0/G1switch 2 |
C00002526
|
| 2548 | GAA, LYAG | glucosidase, alpha; acid (EC:3.2.1.20) |
C00002526
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00002526
|
| 10912 | GADD45G, CR6, DDIT2, GADD45gamma, GRP17 | growth arrest and DNA-damage-inducible, gamma |
C00002526
|
| 8693 | GALNT4, GALNAC-T4, GALNACT4 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) (EC:2.4.1.41) |
C00002526
|
| 8811 | GALR2, GAL2-R, GALNR2, GALR-2 | galanin receptor 2 |
C00002526
|
| 2621 | GAS6, AXLLG, AXSF | growth arrest-specific 6 |
C00002526
|
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible |
C00002526
|
| 2634 | GBP2 | guanylate binding protein 2, interferon-inducible |
C00002526
|
| 9615 | GDA, CYPIN, GUANASE, NEDASIN | guanine deaminase (EC:3.5.4.3) |
C00002526
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002526
|
| 81544 | GDPD5, GDE2, PP1665 | glycerophosphodiester phosphodiesterase domain containing 5 |
C00002526
|
| 2674 | GFRA1, GDNFR, GDNFRA, GFR-ALPHA-1, RET1L, RETL1, TRNR1 | GDNF family receptor alpha 1 |
C00002526
|
| 8836 | GGH, GH | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (EC:3.4.19.9) |
C00002526
|
| 9837 | GINS1, PSF1 | GINS complex subunit 1 (Psf1 homolog) |
C00002526
|
| 51659 | GINS2, HSPC037, PSF2, Pfs2 | GINS complex subunit 2 (Psf2 homolog) |
C00002526
|
| 10755 | GIPC1, C19orf3, GIPC, GLUT1CBP, Hs.6454, IIP-1, NIP, RGS19IP1, SEMCAP, SYNECTIIN, SYNECTIN, TIP-2 | GIPC PDZ domain containing family, member 1 |
C00002526
|
| 2731 | GLDC, GCE, GCSP, HYGN1 | glycine dehydrogenase (decarboxylating) (EC:1.4.4.2) |
C00002526
|
| 2741 | GLRA1, HKPX1, STHE | glycine receptor, alpha 1 |
C00002526
|
| 51022 | GLRX2, CGI-133, GRX2 | glutaredoxin 2 |
C00002526
|
| 2752 | GLUL, GLNS, GS, PIG43, PIG59 | glutamate-ammonia ligase (EC:4.1.1.15 6.3.1.2) |
C00002526
|
| 51053 | GMNN, Gem | geminin, DNA replication inhibitor |
C00002526
|
| 2783 | GNB2 | guanine nucleotide binding protein (G protein), beta polypeptide 2 |
C00002526
|
| 2788 | GNG7 | guanine nucleotide binding protein (G protein), gamma 7 |
C00002526
|
| 2806 | GOT2, KAT4, KATIV, mitAAT | glutamic-oxaloacetic transaminase 2, mitochondrial (EC:2.6.1.1 2.6.1.7) |
C00002526
|
| 2852 | GPER1, CEPR, CMKRL2, DRY12, FEG-1, GPCR-Br, GPER, GPR30, LERGU, LERGU2, LyGPR | G protein-coupled estrogen receptor 1 |
C00002526
|
| 9289 | GPR56, BFPP, TM7LN4, TM7XN1 | G protein-coupled receptor 56 |
C00002526
|
| 9052 | GPRC5A, GPCR5A, RAI3, RAIG1 | G protein-coupled receptor, family C, group 5, member A |
C00002526
|
| 2873 | GPS1, COPS1, CSN1 | G protein pathway suppressor 1 |
C00002526
|
| 26086 | GPSM1, AGS3 | G-protein signaling modulator 1 |
C00002526
|
| 2876 | GPX1, GPXD, GSHPX1 | glutathione peroxidase 1 (EC:1.11.1.9) |
C00002526
|
| 2885 | GRB2, ASH, EGFRBP-GRB2, Grb3-3, MST084, MSTP084, NCKAP2 | growth factor receptor-bound protein 2 |
C00002526
|
| 9687 | GREB1 | growth regulation by estrogen in breast cancer 1 |
C00002526
|
| 2932 | GSK3B | glycogen synthase kinase 3 beta (EC:2.7.11.1 2.7.11.26) |
C00002526
|
| 2936 | GSR | glutathione reductase (EC:1.8.1.7) |
C00002526
|
| 2937 | GSS, GSHS | glutathione synthetase (EC:6.3.2.3) |
C00002526
|
| 2962 | GTF2F1, BTF4, RAP74, TF2F1, TFIIF | general transcription factor IIF, polypeptide 1, 74kDa |
C00002526
|
| 51512 | GTSE1, B99 | G-2 and S-phase expressed 1 |
C00002526
|
| 3001 | GZMA, CTLA3, HFSP | granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) (EC:3.4.21.78) |
C00002526
|
| 3005 | H1F0, H10, H1FV | H1 histone family, member 0 |
C00002526
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00002526
|
| 3015 | H2AFZ, H2A.Z-1, H2A.z, H2A/z, H2AZ | H2A histone family, member Z |
C00002526
|
| 3033 | HADH, HAD, HADH1, HADHSC, HCDH, HHF4, MSCHAD, SCHAD | hydroxyacyl-CoA dehydrogenase (EC:1.1.1.35) |
C00002526
|
| 3036 | HAS1, HAS | hyaluronan synthase 1 (EC:2.4.1.212) |
C00002526
|
| 26959 | HBP1 | HMG-box transcription factor 1 |
C00002526
|
| 54985 | HCFC1R1, HPIP | host cell factor C1 regulator 1 (XPO1 dependent) |
C00002526
|
| 610 | HCN2, BCNG-2, BCNG2, HAC-1 | hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
C00002526
|
| 10870 | HCST, DAP10, KAP10, PIK3AP | hematopoietic cell signal transducer |
C00002526
|
| 63897 | HEATR6, ABC1 | HEAT repeat containing 6 |
C00002526
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00002526
|
| 54626 | HES2, bHLHb40 | hes family bHLH transcription factor 2 |
C00002526
|
| 284004 | HEXDC | hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing (EC:3.2.1.52) |
C00002526
|
| 3091 | HIF1A, HIF-1A, HIF-1alpha, HIF1, HIF1-ALPHA, MOP1, PASD8, bHLHe78 | hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
C00002526
|
| 3009 | HIST1H1B, H1, H1.5, H1B, H1F5, H1s-3 | histone cluster 1, H1b |
C00002526
|
| 3006 | HIST1H1C, H1.2, H1C, H1F2, H1s-1 | histone cluster 1, H1c |
C00002526
|
| 3008 | HIST1H1E, H1.4, H1E, H1F4, H1s-4, dJ221C16.5 | histone cluster 1, H1e |
C00002526
|
| 8335 | HIST1H2AB, H2A/m, H2AFM | histone cluster 1, H2ab |
C00002526
|
| 8334 | HIST1H2AC, H2A/l, H2AFL, dJ221C16.4 | histone cluster 1, H2ac |
C00002526
|
| 8969 | HIST1H2AG, H2A.1b, H2A/p, H2AFP, H2AG, pH2A/f | histone cluster 1, H2ag |
C00002526
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00002526
|
| 8341 | HIST1H2BN, H2B/d, H2BFD | histone cluster 1, H2bn |
C00002526
|
| 8359 | HIST1H4A, H4FA | histone cluster 1, H4a |
C00002526
|
| 8349 | HIST2H2BE, GL105, H2B, H2B.1, H2BFQ, H2BGL105, H2BQ | histone cluster 2, H2be |
C00002526
|
| 8290 | HIST3H3, H3.4, H3/g, H3FT, H3t | histone cluster 3, H3 |
C00002526
|
| 3096 | HIVEP1, CIRIP, CRYBP1, MBP-1, PRDII-BF1, Schnurri-1, ZAS1, ZNF40, ZNF40A | human immunodeficiency virus type I enhancer binding protein 1 |
C00002526
|
| 3099 | HK2, HKII, HXK2 | hexokinase 2 (EC:2.7.1.1) |
C00002526
|
| 3122 | HLA-DRA, HLA-DRA1, MLRW | major histocompatibility complex, class II, DR alpha |
C00002526
|
| 81502 | HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3, PSL3, SPP, SPPL1, dJ324O17.1 | histocompatibility (minor) 13 |
C00002526
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00002526
|
| 3155 | HMGCL, HL | 3-hydroxymethyl-3-methylglutaryl-CoA lyase (EC:4.1.3.4) |
C00002526
|
| 3150 | HMGN1, HMG14 | high mobility group nucleosome binding domain 1 |
C00002526
|
| 3151 | HMGN2, HMG17 | high mobility group nucleosomal binding domain 2 |
C00002526
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00002526
|
| 3181 | HNRNPA2B1, HNRNPA2, HNRNPB1, HNRPA2, HNRPA2B1, HNRPB1, IBMPFD2, RNPA2, SNRPB1 | heterogeneous nuclear ribonucleoprotein A2/B1 |
C00002526
|
| 3183 | HNRNPC, C1, C2, HNRNP, HNRPC, SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) |
C00002526
|
| 221092 | HNRNPUL2, HNRPUL2, SAF-A2 | heterogeneous nuclear ribonucleoprotein U-like 2 |
C00002526
|
| 9455 | HOMER2, ACPD, CPD, HOMER-2, VESL-2 | homer homolog 2 (Drosophila) |
C00002526
|
| 3206 | HOXA10, HOX1, HOX1.8, HOX1H, PL | homeobox A10 |
C00002526
|
| 55806 | HR, ALUNC, AU, HSA277165, MUHH, MUHH1 | hair growth associated |
C00002526
|
| 3291 | HSD11B2, AME, AME1, HSD11K, HSD2, SDR9C3 | hydroxysteroid (11-beta) dehydrogenase 2 (EC:1.1.1.146) |
C00002526
|
| 3292 | HSD17B1, EDH17B2, EDHB17, HSD17, SDR28C1 | hydroxysteroid (17-beta) dehydrogenase 1 (EC:1.1.1.62) |
C00002526
|
| 7923 | HSD17B8, D6S2245E, FABG, FABGL, H2-KE6, HKE6, KE6, RING2, SDR30C1, dJ1033B10.9 | hydroxysteroid (17-beta) dehydrogenase 8 (EC:1.1.1.62 1.1.1.239) |
C00002526
|
| 3306 | HSPA2, HSP70-2, HSP70-3 | heat shock 70kDa protein 2 |
C00002526
|
| 22824 | HSPA4L, APG-1, HSPH3, Osp94 | heat shock 70kDa protein 4-like |
C00002526
|
| 3313 | HSPA9, CSA, GRP-75, GRP75, HSPA9B, MOT, MOT2, MTHSP75, PBP74 | heat shock 70kDa protein 9 (mortalin) |
C00002526
|
| 26353 | HSPB8, CMT2L, DHMN2, E2IG1, H11, HMN2, HMN2A, HSP22 | heat shock 22kDa protein 8 |
C00002526
|
| 81888 | HYI, HT036 | hydroxypyruvate isomerase (putative) (EC:5.3.1.22) |
C00002526
|
| 3397 | ID1, ID, bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
C00002526
|
| 3399 | ID3, HEIR-1, bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
C00002526
|
| 3422 | IDI1, IPP1, IPPI1 | isopentenyl-diphosphate delta isomerase 1 (EC:5.3.3.2) |
C00002526
|
| 389792 | IER5L, bA247A12.2 | immediate early response 5-like |
C00002526
|
| 10410 | IFITM3, 1-8U, DSPA2b, IP15 | interferon induced transmembrane protein 3 |
C00002526
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002526
|
| 282618 | IFNL1, IL-29, IL29 | interferon, lambda 1 |
C00002526
|
| 7866 | IFRD2, IFNRP, SKMc15, SM15 | interferon-related developmental regulator 2 |
C00002526
|
| 3479 | IGF1, IGF-I, IGF1A, IGFI | insulin-like growth factor 1 (somatomedin C) |
C00002526
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00002526
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00002526
|
| 3487 | IGFBP4, BP-4, HT29-IGFBP, IBP4, IGFBP-4 | insulin-like growth factor binding protein 4 |
C00002526
|
| 93185 | IGSF8, CD316, CD81P3, EWI-2, EWI2, KCT-4, LIR-D1, PGRL | immunoglobulin superfamily, member 8 |
C00002526
|
| 3600 | IL15, IL-15 | interleukin 15 |
C00002526
|
| 3552 | IL1A, IL-1A, IL1, IL1-ALPHA, IL1F1 | interleukin 1, alpha |
C00002526
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00002526
|
| 7850 | IL1R2, CD121b, CDw121b, IL-1R-2, IL-1RT-2, IL-1RT2, IL1R2c, IL1RB | interleukin 1 receptor, type II |
C00002526
|
| 3566 | IL4R, CD124, IL-4RA, IL4RA | interleukin 4 receptor |
C00002526
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002526
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002526
|
| 3608 | ILF2, NF45, PRO3063 | interleukin enhancer binding factor 2 |
C00002526
|
| 3609 | ILF3, CBTF, DRBF, DRBP76, MMP4, MPHOSPH4, MPP4, NF-AT-90, NF110, NF110b, NF90, NF90a, NF90b, NFAR, NFAR-1, NFAR2, TCP110, TCP80 | interleukin enhancer binding factor 3, 90kDa |
C00002526
|
| 3615 | IMPDH2, IMPD2, IMPDH-II | IMP (inosine 5'-monophosphate) dehydrogenase 2 (EC:1.1.1.205) |
C00002526
|
| 8821 | INPP4B | inositol polyphosphate-4-phosphatase, type II, 105kDa (EC:3.1.3.66) |
C00002526
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00002526
|
| 9670 | IPO13, IMP13, KAP13, LGL2, RANBP13 | importin 13 |
C00002526
|
| 79711 | IPO4, Imp4 | importin 4 |
C00002526
|
| 10788 | IQGAP2 | IQ motif containing GTPase activating protein 2 |
C00002526
|
| 26145 | IRF2BP1 | interferon regulatory factor 2 binding protein 1 |
C00002526
|
| 3665 | IRF7, IRF-7H, IRF7A, IRF7B, IRF7C, IRF7H | interferon regulatory factor 7 |
C00002526
|
| 8660 | IRS2, IRS-2 | insulin receptor substrate 2 |
C00002526
|
| 9636 | ISG15, G1P2, IFI15, IP17, UCRP, hUCRP | ISG15 ubiquitin-like modifier |
C00002526
|
| 51477 | ISYNA1, INO1, INOS, IPS, IPS_1, IPS-1 | inositol-3-phosphate synthase 1 (EC:5.5.1.4) |
C00002526
|
| 3685 | ITGAV, CD51, MSK8, VNRA, VTNR | integrin, alpha V |
C00002526
|
| 3690 | ITGB3, BDPLT16, BDPLT2, CD61, GP3A, GPIIIa, GT | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
C00002526
|
| 3694 | ITGB6 | integrin, beta 6 |
C00002526
|
| 3705 | ITPK1, ITRPK1 | inositol-tetrakisphosphate 1-kinase (EC:2.7.1.134 2.7.1.159) |
C00002526
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00002526
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00002526
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00002526
|
| 3728 | JUP, ARVD12, CTNNG, DP3, DPIII, PDGB, PKGB | junction plakoglobin |
C00002526
|
| 23254 | KAZN, KAZ | kazrin, periplakin interacting protein |
C00002526
|
| 3754 | KCNF1, IK8, KCNF, KV5.1, kH1 | potassium voltage-gated channel, subfamily F, member 1 |
C00002526
|
| 26251 | KCNG2, KCNF2, KV6.2 | potassium voltage-gated channel, subfamily G, member 2 |
C00002526
|
| 3762 | KCNJ5, CIR, GIRK4, KATP1, KIR3.4, LQT13 | potassium inwardly-rectifying channel, subfamily J, member 5 |
C00002526
|
| 60598 | KCNK15, K2p15.1, KCNK11, KCNK14, KT3.3, TASK-5, TASK5, dJ781B1.1 | potassium channel, subfamily K, member 15 |
C00002526
|
| 23510 | KCTD2 | potassium channel tetramerization domain containing 2 |
C00002526
|
| 222658 | KCTD20, C6orf69, dJ108K11.3 | potassium channel tetramerization domain containing 20 |
C00002526
|
| 10945 | KDELR1, ERD2, ERD2.1, HDEL, PM23 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
C00002526
|
| 9768 | NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | KIAA0101 |
C00002526
|
| 282442 |
C00002526
|
||
| 9851 | KIAA0753 |
C00002526
|
|
| 23285 | KIAA1107 |
C00002526
|
|
| 57648 | KIAA1522 |
C00002526
|
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00002526
|
| 9928 | KIF14 | kinesin family member 14 |
C00002526
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00002526
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00002526
|
| 9493 | KIF23, CHO1, KNSL5, MKLP-1, MKLP1 | kinesin family member 23 |
C00002526
|
| 11004 | KIF2C, KNSL6, MCAK | kinesin family member 2C |
C00002526
|
| 24137 | KIF4A, KIF4, KIF4G1 | kinesin family member 4A |
C00002526
|
| 9365 | KL | klotho (EC:3.2.1.31) |
C00002526
|
| 54800 | KLHL24, DRE1, KRIP6 | kelch-like family member 24 |
C00002526
|
| 3817 | KLK2, KLK2A2, hGK-1, hK2 | kallikrein-related peptidase 2 (EC:3.4.21.35) |
C00002526
|
| 354 | KLK3, APS, KLK2A1, PSA, hK3 | kallikrein-related peptidase 3 (EC:3.4.21.77) |
C00002526
|
| 4297 | KMT2A, ALL-1, CXXC7, HRX, HTRX1, MLL, MLL/GAS7, MLL1A, TET1-MLL, TRX1, WDSTS | lysine (K)-specific methyltransferase 2A (EC:2.1.1.43) |
C00002526
|
| 3838 | KPNA2, IPOA1, QIP2, RCH1, SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
C00002526
|
| 448834 | KPRP, C1orf45 | keratinocyte proline-rich protein |
C00002526
|
| 3845 | KRAS, C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2 | Kirsten rat sarcoma viral oncogene homolog |
C00002526
|
| 3872 | KRT17, K17, PC, PC2, PCHC1 | keratin 17 |
C00002526
|
| 3875 | KRT18, CYK18, K18 | keratin 18 |
C00002526
|
| 3880 | KRT19, CK19, K19, K1CS | keratin 19 |
C00002526
|
| 25984 | KRT23, CK23, HAIK1, K23 | keratin 23 (histone deacetylase inducible) |
C00002526
|
| 3885 | KRT34, HA4, Ha-4, KRTHA4, hHa4 | keratin 34 |
C00002526
|
| 3852 | KRT5, CK5, DDD, DDD1, EBS2, K5, KRT5A | keratin 5 |
C00002526
|
| 3856 | KRT8, CARD2, CK-8, CK8, CYK8, K2C8, K8, KO | keratin 8 |
C00002526
|
| 90133 |
C00002526
|
||
| 728279 | KRTAP2-2, KAP2.2 | keratin associated protein 2-2 |
C00002526
|
| 85294 | KRTAP2-4, KAP2.1B, KAP2.4, KRTAP2.4 | keratin associated protein 2-4 |
C00002526
|
| 3897 | L1CAM, CAML1, CD171, HSAS, HSAS1, MASA, MIC5, N-CAM-L1, N-CAML1, NCAM-L1, S10, SPG1 | L1 cell adhesion molecule |
C00002526
|
| 3915 | LAMC1, LAMB2 | laminin, gamma 1 (formerly LAMB2) |
C00002526
|
| 3920 | LAMP2, CD107b, LAMP-2, LAMPB, LGP110 | lysosomal-associated membrane protein 2 |
C00002526
|
| 3945 | LDHB, LDH-B, LDH-H, LDHBD, TRG-5 | lactate dehydrogenase B (EC:1.1.1.27) |
C00002526
|
| 7044 | LEFTY2, EBAF, LEFTA, LEFTYA, TGFB4 | left-right determination factor 2 |
C00002526
|
| 93273 | LEMD1, CT50, LEMP-1 | LEM domain containing 1 |
C00002526
|
| 3952 | LEP, LEPD, OB, OBS | leptin |
C00002526
|
| 3955 | LFNG, SCDO3 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (EC:2.4.1.222) |
C00002526
|
| 3956 | LGALS1, GAL1, GBP | lectin, galactoside-binding, soluble, 1 |
C00002526
|
| 3963 | LGALS7, GAL7, LGALS7A | lectin, galactoside-binding, soluble, 7 |
C00002526
|
| 3980 | LIG3, LIG2 | ligase III, DNA, ATP-dependent (EC:6.5.1.1) |
C00002526
|
| 4000 | LMNA, CDCD1, CDDC, CMD1A, CMT2B1, EMD2, FPL, FPLD, FPLD2, HGPS, IDC, LDP1, LFP, LGMD1B, LMN1, LMNC, LMNL1, PRO1 | lamin A/C |
C00002526
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00002526
|
| 84823 | LMNB2, LAMB2, LMN2 | lamin B2 |
C00002526
|
| 9170 | LPAR2, EDG-4, EDG4, LPA-2, LPA2 | lysophosphatidic acid receptor 2 |
C00002526
|
| 4026 | LPP | LIM domain containing preferred translocation partner in lipoma |
C00002526
|
| 64748 | LPPR2, PRG4 | lipid phosphate phosphatase-related protein type 2 (EC:3.1.3.4) |
C00002526
|
| 91355 | LRP5L | low density lipoprotein receptor-related protein 5-like |
C00002526
|
| 7804 | LRP8, APOER2, HSZ75190, LRP-8, MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
C00002526
|
| 131578 | LRRC15, LIB | leucine rich repeat containing 15 |
C00002526
|
| 4057 | LTF, GIG12, HLF2, LF | lactotransferrin |
C00002526
|
| 56925 | LXN, ECI, TCI | latexin |
C00002526
|
| 90624 | LYRM7, C5orf31, MZM1L | LYR motif containing 7 |
C00002526
|
| 8379 | MAD1L1, MAD1, PIG9, TP53I9, TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) |
C00002526
|
| 8567 | MADD, DENN, IG20, RAB3GEP | MAP-kinase activating death domain |
C00002526
|
| 4094 | MAF, CCA4, c-MAF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
C00002526
|
| 9935 | MAFB, KRML, MCTO | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
C00002526
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00002526
|
| 28986 | MAGEH1, APR-1, APR1, MAGEH | melanoma antigen family H, 1 |
C00002526
|
| 4128 | MAOA, MAO-A | monoamine oxidase A (EC:1.4.3.4) |
C00002526
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00002526
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00002526
|
| 5606 | MAP2K3, MAPKK3, MEK3, MKK3, PRKMK3, SAPKK-2, SAPKK2 | mitogen-activated protein kinase kinase 3 (EC:2.7.12.2) |
C00002526
|
| 4134 | MAP4 | microtubule-associated protein 4 |
C00002526
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002526
|
| 5600 | MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2, p38Beta | mitogen-activated protein kinase 11 (EC:2.7.11.24) |
C00002526
|
| 5603 | MAPK13, MAPK_13, MAPK-13, PRKM13, SAPK4, p38delta | mitogen-activated protein kinase 13 (EC:2.7.11.24) |
C00002526
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002526
|
| 84930 | MASTL, GREATWALL, GW, GWL, MAST-L, RP11-85G18.2, THC2, hGWL | microtubule associated serine/threonine kinase-like (EC:2.7.11.1) |
C00002526
|
| 4145 | MATK, CHK, CTK, HHYLTK, HYL, HYLTK, Lsk | megakaryocyte-associated tyrosine kinase (EC:2.7.10.2) |
C00002526
|
| 9782 | MATR3, MPD2, VCPDM | matrin 3 |
C00002526
|
| 4150 | MAZ, PUR1, Pur-1, SAF-1, SAF-2, SAF-3, ZF87, ZNF801, Zif87 | MYC-associated zinc finger protein (purine-binding transcription factor) |
C00002526
|
| 8932 | MBD2, DMTase, NY-CO-41 | methyl-CpG binding domain protein 2 |
C00002526
|
| 4157 | MC1R, CMM5, MSH-R, SHEP2 | melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
C00002526
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00002526
|
| 4171 | MCM2, BM28, CCNL1, CDCL1, D3S3194, MITOTIN, cdc19 | minichromosome maintenance complex component 2 (EC:3.6.4.12) |
C00002526
|
| 4175 | MCM6, MCG40308, Mis5, P105MCM | minichromosome maintenance complex component 6 (EC:3.6.4.12) |
C00002526
|
| 4176 | MCM7, CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146 | minichromosome maintenance complex component 7 (EC:3.6.4.12) |
C00002526
|
| 9656 | MDC1, NFBD1 | mediator of DNA-damage checkpoint 1 |
C00002526
|
| 4192 | MDK, ARAP, MK, NEGF2 | midkine (neurite growth-promoting factor 2) |
C00002526
|
| 4199 | ME1, HUMNDME, MES | malic enzyme 1, NADP(+)-dependent, cytosolic (EC:1.1.1.40) |
C00002526
|
| 4204 | MECP2, AUTSX3, MRX16, MRX79, MRXS13, MRXSL, PPMX, RS, RTS, RTT | methyl CpG binding protein 2 (Rett syndrome) |
C00002526
|
| 9282 | MED14, CRSP150, CRSP2, CSRP, CXorf4, DRIP150, EXLM1, RGR1, TRAP170 | mediator complex subunit 14 |
C00002526
|
| 9477 | MED20, PRO0213, TRFP | mediator complex subunit 20 |
C00002526
|
| 9833 | MELK, HPK38 | maternal embryonic leucine zipper kinase (EC:2.7.11.1 2.7.10.2) |
C00002526
|
| 4221 | MEN1, MEAI, SCG2 | multiple endocrine neoplasia I |
C00002526
|
| 4222 | MEOX1, KFS2, MOX1 | mesenchyme homeobox 1 |
C00002526
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00002526
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00002526
|
| 4240 | MFGE8, BA46, EDIL1, HMFG, HsT19888, MFG-E8, MFGM, OAcGD3S, SED1, SPAG10, hP47 | milk fat globule-EGF factor 8 protein |
C00002526
|
| 9927 | MFN2, CMT2A, CMT2A2, CPRP1, HSG, MARF | mitofusin 2 |
C00002526
|
| 4242 | MFNG | MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (EC:2.4.1.222) |
C00002526
|
| 4255 | MGMT | O-6-methylguanine-DNA methyltransferase (EC:2.1.1.63) |
C00002526
|
| 4256 | MGP, MGLAP, NTI | matrix Gla protein |
C00002526
|
| 574449 | MIR492, MIRN492, hsa-mir-492 | microRNA 492 |
C00002526
|
| 724033 | MIR663A, MIR663, MIRN663, hsa-mir-663, hsa-mir-663a | microRNA 663a |
C00002526
|
| 4291 | MLF1 | myeloid leukemia factor 1 |
C00002526
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00002526
|
| 197259 | MLKL | mixed lineage kinase domain-like |
C00002526
|
| 4301 | MLLT4, AF6, RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
C00002526
|
| 79083 | MLPH, SLAC2-A | melanophilin |
C00002526
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00002526
|
| 4323 | MMP14, MMP-14, MMP-X1, MT-MMP, MT-MMP_1, MT1-MMP, MT1MMP, MTMMP1, WNCHRS | matrix metallopeptidase 14 (membrane-inserted) (EC:3.4.24.80) |
C00002526
|
| 4324 | MMP15, MT2-MMP, MTMMP2, SMCP-2 | matrix metallopeptidase 15 (membrane-inserted) (EC:3.4.24.-) |
C00002526
|
| 4325 | MMP16, C8orf57, MMP-X2, MT-MMP2, MT-MMP3, MT3-MMP | matrix metallopeptidase 16 (membrane-inserted) (EC:3.4.24.-) |
C00002526
|
| 4326 | MMP17, MT4-MMP | matrix metallopeptidase 17 (membrane-inserted) (EC:3.4.24.-) |
C00002526
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00002526
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00002526
|
| 4331 | MNAT1, CAP35, MAT1, RNF66, TFB3 | MNAT CDK-activating kinase assembly factor 1 |
C00002526
|
| 4335 | MNT, MAD6, MXD6, ROX, bHLHd3 | MAX network transcriptional repressor |
C00002526
|
| 7841 | MOGS, CDG2B, CWH41, DER7, GCS1 | mannosyl-oligosaccharide glucosidase (EC:3.2.1.106) |
C00002526
|
| 10198 | MPHOSPH9, MPP-9, MPP9 | M-phase phosphoprotein 9 |
C00002526
|
| 84954 | MPND | MPN domain containing |
C00002526
|
| 55052 | MRPL20, L20mt, MRP-L20 | mitochondrial ribosomal protein L20 |
C00002526
|
| 740 | MRPL49, C11orf4, L49mt, MRP-L49, NOF, NOF1 | mitochondrial ribosomal protein L49 |
C00002526
|
| 124540 | MSI2, MSI2H | musashi RNA-binding protein 2 |
C00002526
|
| 4477 | MSMB, HPC13, IGBF, MSP, MSPB, PN44, PRPS, PSP, PSP-94, PSP57, PSP94 | microseminoprotein, beta- |
C00002526
|
| 4501 | MT1X, MT-1l, MT1 | metallothionein 1X |
C00002526
|
| 4502 | MT2A, MT2 | metallothionein 2A |
C00002526
|
| 4522 | MTHFD1, MTHFC, MTHFD | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase (EC:1.5.1.5 6.3.4.3 3.5.4.9) |
C00002526
|
| 4582 | MUC1, CA_15-3, CD227, EMA, H23AG, KL-6, MAM6, MCKD1, MUC-1, MUC-1/SEC, MUC-1/X, MUC1/ZD, PEM, PEMT, PUM | mucin 1, cell surface associated |
C00002526
|
| 4597 | MVD, FP17780, MPD | mevalonate (diphospho) decarboxylase (EC:4.1.1.33) |
C00002526
|
| 4599 | MX1, IFI-78K, IFI78, MX, MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
C00002526
|
| 10608 | MXD4, MAD4, MST149, MSTP149, bHLHc12 | MAX dimerization protein 4 |
C00002526
|
| 439921 | MXRA7, PS1TP1, TMAP1 | matrix-remodelling associated 7 |
C00002526
|
| 4602 | MYB, Cmyb, c-myb, c-myb_CDS, efg | v-myb avian myeloblastosis viral oncogene homolog |
C00002526
|
| 10514 | MYBBP1A, P160, PAP2 | MYB binding protein (P160) 1a |
C00002526
|
| 4605 | MYBL2, B-MYB, BMYB | v-myb avian myeloblastosis viral oncogene homolog-like 2 |
C00002526
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00002526
|
| 4628 | MYH10, NMMHC-IIB, NMMHCB | myosin, heavy chain 10, non-muscle |
C00002526
|
| 79784 | MYH14, DFNA4, DFNA4A, MHC16, MYH17, NMHC_II-C, NMHC-II-C, PNMHH, myosin | myosin, heavy chain 14, non-muscle (EC:3.6.4.1) |
C00002526
|
| 4624 | MYH6, ASD3, CMD1EE, CMH14, MYHC, MYHCA, SSS3, alpha-MHC | myosin, heavy chain 6, cardiac muscle, alpha |
C00002526
|
| 4627 | MYH9, BDPLT6, DFNA17, EPSTS, FTNS, MHA, NMHC-II-A, NMMHC-IIA, NMMHCA | myosin, heavy chain 9, non-muscle |
C00002526
|
| 103910 | MYL12B, MLC-B, MRLC2 | myosin, light chain 12B, regulatory |
C00002526
|
| 4637 | MYL6, ESMLC, LC17, LC17-GI, LC17-NM, LC17A, LC17B, MLC-3, MLC1SM, MLC3NM, MLC3SM | myosin, light chain 6, alkali, smooth muscle and non-muscle |
C00002526
|
| 29116 | MYLIP, IDOL, MIR | myosin regulatory light chain interacting protein (EC:6.3.2.-) |
C00002526
|
| 51168 | MYO15A, DFNB3, MYO15 | myosin XVA |
C00002526
|
| 399687 | MYO18A, MYSPDZ, SPR210 | myosin XVIIIA |
C00002526
|
| 27163 | NAAA, ASAHL, PLT | N-acylethanolamine acid amidase (EC:3.5.1.-) |
C00002526
|
| 4665 | NAB2, MADER | NGFI-A binding protein 2 (EGR1 binding protein 2) |
C00002526
|
| 4676 | NAP1L4, NAP1L4b, NAP2, NAP2L, hNAP2 | nucleosome assembly protein 1-like 4 |
C00002526
|
| 8775 | NAPA, SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha |
C00002526
|
| 26502 | NARF, IOP2 | nuclear prelamin A recognition factor |
C00002526
|
| 4678 | NASP, FLB7527, PRO1999 | nuclear autoantigenic sperm protein (histone-binding) |
C00002526
|
| 339983 | NAT8L, CML3, NACED, NAT8-LIKE | N-acetyltransferase 8-like (GCN5-related, putative) (EC:2.3.1.17) |
C00002526
|
| 4683 | NBN, AT-V1, AT-V2, ATV, NBS, NBS1, P95 | nibrin |
C00002526
|
| 9918 | NCAPD2, CAP-D2, CNAP1, hCAP-D2 | non-SMC condensin I complex, subunit D2 |
C00002526
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00002526
|
| 400746 | NCMAP, C1orf130, MP11 | noncompact myelin associated protein |
C00002526
|
| 8648 | NCOA1, F-SRC-1, KAT13A, RIP160, SRC1, bHLHe42, bHLHe74 | nuclear receptor coactivator 1 (EC:2.3.1.48) |
C00002526
|
| 10499 | NCOA2, GRIP1, KAT13C, NCoA-2, SRC2, TIF2, bHLHe75 | nuclear receptor coactivator 2 |
C00002526
|
| 8202 | NCOA3, ACTR, AIB-1, AIB1, CAGH16, CTG26, KAT13B, RAC3, SRC-3, SRC3, TNRC14, TNRC16, TRAM-1, bHLHe42, pCIP | nuclear receptor coactivator 3 (EC:2.3.1.48) |
C00002526
|
| 8031 | NCOA4, ARA70, ELE1, PTC3, RFG | nuclear receptor coactivator 4 |
C00002526
|
| 10403 | NDC80, HEC, HEC1, HsHec1, KNTC2, TID3, hsNDC80 | NDC80 kinetochore complex component |
C00002526
|
| 4707 | NDUFB1, CI-MNLL, CI-SGDH, MNLL | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
C00002526
|
| 4739 | NEDD9, CAS-L, CAS2, CASL, CASS2, HEF1 | neural precursor cell expressed, developmentally down-regulated 9 |
C00002526
|
| 252969 | NEIL2, NEH2, NEI2 | nei endonuclease VIII-like 2 (E. coli) (EC:4.2.99.18) |
C00002526
|
| 4753 | NELL2, NRP2 | NEL-like 2 (chicken) |
C00002526
|
| 10725 | NFAT5, NF-AT5, NFATL1, NFATZ, OREBP, TONEBP | nuclear factor of activated T-cells 5, tonicity-responsive |
C00002526
|
| 4790 | NFKB1, EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
C00002526
|
| 4791 | NFKB2, H2TF1, LYT-10, LYT10, NF-kB2, p105, p52 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
C00002526
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00002526
|
| 9054 | NFS1, IscS, NIFS | NFS1 cysteine desulfurase (EC:2.8.1.7) |
C00002526
|
| 374354 | NHLRC2 | NHL repeat containing 2 |
C00002526
|
| 4814 | NINJ1, NIN1, NINJURIN | ninjurin 1 |
C00002526
|
| 348938 | NIPAL4, ARCI6, ICHTHYIN, ICHYN | NIPA-like domain containing 4 |
C00002526
|
| 25836 | NIPBL, CDLS, CDLS1, IDN3, IDN3-B, Scc2 | Nipped-B homolog (Drosophila) |
C00002526
|
| 4824 | NKX3-1, BAPX2, NKX3, NKX3.1, NKX3A | NK3 homeobox 1 |
C00002526
|
| 57486 | NLN, AGTBP, EP24.16, MEP, MOP | neurolysin (metallopeptidase M3 family) (EC:3.4.24.16) |
C00002526
|
| 4836 | NMT1, NMT | N-myristoyltransferase 1 (EC:2.3.1.97) |
C00002526
|
| 65083 | NOL6, NRAP, UTP22, bA311H10.1 | nucleolar protein 6 (RNA-associated) |
C00002526
|
| 4839 | NOP2, NOL1, NOP120, NSUN1, p120 | NOP2 nucleolar protein |
C00002526
|
| 4843 | NOS2, HEP-NOS, INOS, NOS, NOS2A | nitric oxide synthase 2, inducible (EC:1.14.13.39) |
C00002526
|
| 404036 |
C00002526
|
||
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00002526
|
| 10577 | NPC2, EDDM1, HE1 | Niemann-Pick disease, type C2 |
C00002526
|
| 9284 | NPIPA1, NPIP, morpheus | nuclear pore complex interacting protein family, member A1 |
C00002526
|
| 23117 | NPIPB3, KIAA0220L, NPIPB5, NPIPL3 | nuclear pore complex interacting protein family, member B3 |
C00002526
|
| 4869 | NPM1, B23, NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
C00002526
|
| 4880 | NPPC, CNP, CNP2 | natriuretic peptide C |
C00002526
|
| 4881 | NPR1, ANPRA, ANPa, GUC2A, GUCY2A, NPRA | natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) (EC:4.6.1.2) |
C00002526
|
| 283869 | NPW, L8, L8C, PPL8, PPNPW | neuropeptide W |
C00002526
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00002526
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00002526
|
| 7182 | NR2C2, TAK1, TR2R1, TR4, hTAK1 | nuclear receptor subfamily 2, group C, member 2 |
C00002526
|
| 3164 | NR4A1, GFRP1, HMR, N10, NAK-1, NGFIB, NP10, NUR77, TR3 | nuclear receptor subfamily 4, group A, member 1 |
C00002526
|
| 441478 | NRARP | NOTCH-regulated ankyrin repeat protein |
C00002526
|
| 29959 | NRBP1, BCON3, MADM, MUDPNP, NRBP | nuclear receptor binding protein 1 |
C00002526
|
| 8204 | NRIP1, RIP140 | nuclear receptor interacting protein 1 |
C00002526
|
| 56675 | NRIP3, C11orf14, NY-SAR-105 | nuclear receptor interacting protein 3 |
C00002526
|
| 8829 | NRP1, BDCA4, CD304, NP1, NRP, VEGF165R | neuropilin 1 |
C00002526
|
| 8828 | NRP2, NP2, NPN2, PRO2714, VEGF165R2 | neuropilin 2 |
C00002526
|
| 4902 | NRTN, NTN | neurturin |
C00002526
|
| 4926 | NUMA1, NUMA | nuclear mitotic apparatus protein 1 |
C00002526
|
| 158046 | NXNL2, C9orf121, RDCVF2 | nucleoredoxin-like 2 (EC:1.8.1.8) |
C00002526
|
| 100506658 | OCLN, BLCPMG | occludin (EC:2.1.1.67) |
C00002526
|
| 11054 | OGFR | opioid growth factor receptor |
C00002526
|
| 10439 | OLFM1, AMY, NOE1, NOELIN1, OlfA | olfactomedin 1 |
C00002526
|
| 56944 | OLFML3, HNOEL-iso, OLF44 | olfactomedin-like 3 |
C00002526
|
| 29948 | OSGIN1, BDGI, OKL38 | oxidative stress induced growth inhibitor 1 |
C00002526
|
| 78990 | OTUB2, C14orf137, OTB2, OTU2 | OTU domain, ubiquitin aldehyde binding 2 (EC:3.4.19.12) |
C00002526
|
| 5032 | P2RY11, P2Y11 | purinergic receptor P2Y, G-protein coupled, 11 |
C00002526
|
| 5036 | PA2G4, EBP1, HG4-1, p38-2G4 | proliferation-associated 2G4, 38kDa |
C00002526
|
| 5064 | PALM | paralemmin |
C00002526
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00002526
|
| 55872 | PBK, CT84, Nori-3, SPK, TOPK | PDZ binding kinase (EC:2.7.12.2) |
C00002526
|
| 56133 | PCDHB2, PCDH-BETA2 | protocadherin beta 2 |
C00002526
|
| 56130 | PCDHB6, PCDH-BETA6 | protocadherin beta 6 |
C00002526
|
| 5106 | PCK2, PEPCK, PEPCK-M, PEPCK2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) (EC:4.1.1.32) |
C00002526
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00002526
|
| 5121 | PCP4, PEP-19 | Purkinje cell protein 4 |
C00002526
|
| 22984 | PDCD11, ALG-4, ALG4, NFBP, RRP5 | programmed cell death 11 |
C00002526
|
| 27250 | PDCD4, H731 | programmed cell death 4 (neoplastic transformation inhibitor) |
C00002526
|
| 10015 | PDCD6IP, AIP1, ALIX, DRIP4, HP95 | programmed cell death 6 interacting protein |
C00002526
|
| 10081 | PDCD7, ES18, HES18 | programmed cell death 7 |
C00002526
|
| 100135774 |
C00002526
|
||
| 5159 | PDGFRB, CD140B, IBGC4, IMF1, JTK12, PDGFR, PDGFR-1, PDGFR1 | platelet-derived growth factor receptor, beta polypeptide (EC:2.7.10.1) |
C00002526
|
| 10954 | PDIA5, PDIR | protein disulfide isomerase family A, member 5 (EC:5.3.4.1) |
C00002526
|
| 5163 | PDK1 | pyruvate dehydrogenase kinase, isozyme 1 (EC:2.7.11.2) |
C00002526
|
| 64236 | PDLIM2, MYSTIQUE, SLIM | PDZ and LIM domain 2 (mystique) |
C00002526
|
| 5174 | PDZK1, CAP70, CLAMP, NHERF-3, NHERF3, PDZD1 | PDZ domain containing 1 |
C00002526
|
| 23089 | PEG10, EDR, HB-1, MEF3L, Mar2, Mart2, RGAG3 | paternally expressed 10 |
C00002526
|
| 192111 | PGAM5, BXLBV68 | phosphoglycerate mutase family member 5 (EC:3.1.3.16) |
C00002526
|
| 54858 | PGPEP1, PGP, PGP-I, PGPI, Pcp | pyroglutamyl-peptidase I (EC:3.4.19.3) |
C00002526
|
| 5241 | PGR, NR3C3, PR | progesterone receptor |
C00002526
|
| 9678 | PHF14 | PHD finger protein 14 |
C00002526
|
| 7262 | PHLDA2, BRW1C, BWR1C, HLDA2, IPL, TSSC3 | pleckstrin homology-like domain, family A, member 2 |
C00002526
|
| 5296 | PIK3R2, MPPH, P85B, p85, p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
C00002526
|
| 8503 | PIK3R3, p55, p55-GAMMA | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
C00002526
|
| 29990 | PILRB, FDFACT1, FDFACT2 | paired immunoglobin-like type 2 receptor beta |
C00002526
|
| 5292 | PIM1, PIM | pim-1 oncogene (EC:2.7.11.1) |
C00002526
|
| 5304 | PIP, GCDFP-15, GCDFP15, GPIP4 | prolactin-induced protein |
C00002526
|
| 51268 | PIPOX, LPIPOX | pipecolic acid oxidase (EC:1.5.3.1 1.5.3.7) |
C00002526
|
| 26207 | PITPNC1, M-RDGB-beta, MRDGBbeta, RDGB-BETA, RDGBB, RDGBB1 | phosphatidylinositol transfer protein, cytoplasmic 1 |
C00002526
|
| 5320 | PLA2G2A, MOM1, PLA2, PLA2B, PLA2L, PLA2S, PLAS1, sPLA2 | phospholipase A2, group IIA (platelets, synovial fluid) (EC:3.1.1.4) |
C00002526
|
| 5327 | PLAT, T-PA, TPA | plasminogen activator, tissue (EC:3.4.21.68) |
C00002526
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00002526
|
| 23236 | PLCB1, EIEE12, PI-PLC, PLC-154, PLC-I, PLC154, PLCB1A, PLCB1B | phospholipase C, beta 1 (phosphoinositide-specific) (EC:3.1.4.11) |
C00002526
|
| 5339 | PLEC, EBS1, EBSO, HD1, LGMD2Q, PCN, PLEC1, PLEC1b, PLTN | plectin |
C00002526
|
| 10769 | PLK2, SNK, hPlk2, hSNK | polo-like kinase 2 (EC:2.7.11.21) |
C00002526
|
| 10733 | PLK4, SAK, STK18 | polo-like kinase 4 (EC:2.7.11.21) |
C00002526
|
| 5351 | PLOD1, EDS6, LH, LH1, LLH, PLOD | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 (EC:1.14.11.4) |
C00002526
|
| 8985 | PLOD3, LH3 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (EC:1.14.11.4) |
C00002526
|
| 5354 | PLP1, GPM6C, HLD1, MMPL, PLP, PLP/DM20, PMD, SPG2 | proteolipid protein 1 |
C00002526
|
| 5364 | PLXNB1, PLEXIN-B1, PLXN5, SEP | plexin B1 |
C00002526
|
| 56937 | PMEPA1, STAG1, TMEPAI | prostate transmembrane protein, androgen induced 1 |
C00002526
|
| 25953 | PNKD, BRP17, DYT8, FPD1, KIPP1184, MR-1, MR1, PDC, PKND1, TAHCCP2 | paroxysmal nonkinesigenic dyskinesia (EC:3.1.2.6) |
C00002526
|
| 5411 | PNN, DRS, DRSP, SDK3, memA | pinin, desmosome associated protein |
C00002526
|
| 10957 | PNRC1, B4-2, PNAS-145, PROL2, PRR2 | proline-rich nuclear receptor coactivator 1 |
C00002526
|
| 23509 | POFUT1, DDD2, FUT12, O-FUT, O-Fuc-T, O-FucT-1, OFUCT1 | protein O-fucosyltransferase 1 (EC:2.4.1.221) |
C00002526
|
| 57804 | POLD4, POLDS, p12 | polymerase (DNA-directed), delta 4, accessory subunit |
C00002526
|
| 5432 | POLR2C, RPB3, RPB31, hRPB33, hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa (EC:2.7.7.6) |
C00002526
|
| 5442 | POLRMT, APOLMT, MTRNAP, MTRPOL, h-mtRPOL | polymerase (RNA) mitochondrial (DNA directed) (EC:2.7.7.6) |
C00002526
|
| 5443 | POMC, ACTH, CLIP, LPH, MSH, NPP, POC | proopiomelanocortin |
C00002526
|
| 5458 | POU4F2, BRN3.2, BRN3B, Brn-3b | POU class 4 homeobox 2 |
C00002526
|
| 56342 | PPAN, BXDC3, SSF, SSF-1, SSF1, SSF2 | peter pan homolog (Drosophila) |
C00002526
|
| 8613 | PPAP2B, Dri42, LPP3, PAP2B, VCIP | phosphatidic acid phosphatase type 2B (EC:3.1.3.4) |
C00002526
|
| 8612 | PPAP2C, LPP2, PAP-2c, PAP2-g | phosphatidic acid phosphatase type 2C (EC:3.1.3.4) |
C00002526
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00002526
|
| 133522 | PPARGC1B, ERRL1, PERC, PGC-1(beta), PGC1B | peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
C00002526
|
| 8496 | PPFIBP1, L2, SGT2, hSGT2, hSgt2p | PTPRF interacting protein, binding protein 1 (liprin beta 1) |
C00002526
|
| 5493 | PPL | periplakin |
C00002526
|
| 333926 | PPM1J, PP2CZ, PP2Czeta, PPP2CZ | protein phosphatase, Mg2+/Mn2+ dependent, 1J (EC:3.1.3.16) |
C00002526
|
| 5514 | PPP1R10, CAT53, FB19, PNUTS, PP1R10, R111, p99 | protein phosphatase 1, regulatory subunit 10 |
C00002526
|
| 81706 | PPP1R14C, CPI17-like, KEPI, NY-BR-81 | protein phosphatase 1, regulatory (inhibitor) subunit 14C |
C00002526
|
| 84152 | PPP1R1B, DARPP-32, DARPP32 | protein phosphatase 1, regulatory (inhibitor) subunit 1B (EC:3.1.3.16) |
C00002526
|
| 5521 | PPP2R2B, B55BETA, PP2AB55BETA, PP2ABBETA, PP2APR55B, PP2APR55BETA, PR2AB55BETA, PR2ABBETA, PR2APR55BETA, PR52B, PR55-BETA, PR55BETA, SCA12 | protein phosphatase 2, regulatory subunit B, beta (EC:3.1.3.16) |
C00002526
|
| 5522 | PPP2R2C, B55-GAMMA, IMYPNO, IMYPNO1, PR52, PR55G | protein phosphatase 2, regulatory subunit B, gamma (EC:3.1.3.16) |
C00002526
|
| 11230 | PRAF2 | PRA1 domain family, member 2 |
C00002526
|
| 9055 | PRC1, ASE1 | protein regulator of cytokinesis 1 |
C00002526
|
| 5546 | PRCC, RCCP1, TPRC | papillary renal cell carcinoma (translocation-associated) |
C00002526
|
| 57580 | PREX1, P-REX1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
C00002526
|
| 5557 | PRIM1, p49 | primase, DNA, polypeptide 1 (49kDa) (EC:2.7.7.7) |
C00002526
|
| 5562 | PRKAA1, AMPK, AMPKa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit (EC:2.7.11.1 2.7.11.26 2.7.11.27 2.7.11.31) |
C00002526
|
| 5576 | PRKAR2A, PKR2, PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha (EC:2.7.11.1) |
C00002526
|
| 5578 | PRKCA, AAG6, PKC-alpha, PKCA, PRKACA | protein kinase C, alpha (EC:2.7.11.13) |
C00002526
|
| 5580 | PRKCD, MAY1, PKCD, nPKC-delta | protein kinase C, delta (EC:2.7.10.2 2.7.11.13) |
C00002526
|
| 3276 | PRMT1, ANM1, HCP1, HRMT1L2, IR1B4 | protein arginine methyltransferase 1 (EC:2.1.1.125) |
C00002526
|
| 5621 | PRNP, ASCR, AltPrP, CD230, CJD, GSS, KURU, PRIP, PrP, PrP27-30, PrP33-35C, PrPc, p27-30 | prion protein |
C00002526
|
| 5627 | PROS1, PROS, PS21, PS22, PS23, PS24, PS25, PSA, THPH5, THPH6 | protein S (alpha) |
C00002526
|
| 10594 | PRPF8, HPRP8, PRP8, PRPC8, RP13, SNRNP220 | pre-mRNA processing factor 8 |
C00002526
|
| 5644 | PRSS1, TRP1, TRY1, TRY4, TRYP1 | protease, serine, 1 (trypsin 1) (EC:3.4.21.4) |
C00002526
|
| 11098 | PRSS23, SIG13, SPUVE, ZSIG13 | protease, serine, 23 |
C00002526
|
| 5652 | PRSS8, CAP1, PROSTASIN | protease, serine, 8 |
C00002526
|
| 29968 | PSAT1, EPIP, PSA, PSAT | phosphoserine aminotransferase 1 (EC:2.6.1.52) |
C00002526
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00002526
|
| 5731 | PTGER1, EP1 | prostaglandin E receptor 1 (subtype EP1), 42kDa |
C00002526
|
| 5733 | PTGER3, EP3, EP3-I, EP3-II, EP3-III, EP3-IV, EP3e, PGE2-R | prostaglandin E receptor 3 (subtype EP3) |
C00002526
|
| 145482 | PTGR2, PGR2, ZADH1 | prostaglandin reductase 2 (EC:1.3.1.48) |
C00002526
|
| 5742 | PTGS1, COX1, COX3, PCOX1, PES-1, PGG/HS, PGHS-1, PGHS1, PHS1, PTGHS | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002526
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002526
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00002526
|
| 5763 | PTMS, ParaT | parathymosin |
C00002526
|
| 80324 | PUS1, MLASA1 | pseudouridylate synthase 1 (EC:5.4.99.12) |
C00002526
|
| 5822 | PWP2, EHOC-17, PWP2H, UTP1 | PWP2 periodic tryptophan protein homolog (yeast) |
C00002526
|
| 5834 | PYGB, GPBB | phosphorylase, glycogen; brain (EC:2.4.1.1) |
C00002526
|
| 5858 | PZP, CPAMD6 | pregnancy-zone protein |
C00002526
|
| 54870 | QRICH1 | glutamine-rich 1 |
C00002526
|
| 5768 | QSOX1, Q6, QSCN6 | quiescin Q6 sulfhydryl oxidase 1 (EC:1.8.3.2) |
C00002526
|
| 169714 | QSOX2, QSCN6L1, SOXN | quiescin Q6 sulfhydryl oxidase 2 (EC:1.8.3.2) |
C00002526
|
| 9230 | RAB11B, H-YPT3 | RAB11B, member RAS oncogene family |
C00002526
|
| 22931 | RAB18, RAB18LI1, WARBM3 | RAB18, member RAS oncogene family |
C00002526
|
| 10244 | RABEPK, RAB9P40, bA65N13.1, p40 | Rab9 effector protein with kelch motifs |
C00002526
|
| 29127 | RACGAP1, CYK4, HsCYK-4, ID-GAP, MgcRacGAP | Rac GTPase activating protein 1 |
C00002526
|
| 5886 | RAD23A, HHR23A, HR23A | RAD23 homolog A (S. cerevisiae) |
C00002526
|
| 5888 | RAD51, BRCC5, HRAD51, HsRad51, HsT16930, MRMV2, RAD51A, RECA | RAD51 recombinase |
C00002526
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00002526
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00002526
|
| 5894 | RAF1, CRAF, NS5, Raf-1, c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 (EC:2.7.11.1) |
C00002526
|
| 5905 | RANGAP1, Fug1, RANGAP, SD | Ran GTPase activating protein 1 |
C00002526
|
| 51195 | RAPGEFL1, Link-GEFII | Rap guanine nucleotide exchange factor (GEF)-like 1 |
C00002526
|
| 5920 | RARRES3, HRASLS4, HRSL4, PLA1/2-3, RIG1, TIG3 | retinoic acid receptor responder (tazarotene induced) 3 |
C00002526
|
| 22821 | RASA3, GAP1IP4BP, GAPIII | RAS p21 protein activator 3 |
C00002526
|
| 153020 | RASGEF1B, GPIG4 | RasGEF domain family, member 1B |
C00002526
|
| 10125 | RASGRP1, CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
C00002526
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00002526
|
| 5932 | RBBP8, COM1, CTIP, JWDS, RIM, SAE2, SCKL2 | retinoblastoma binding protein 8 |
C00002526
|
| 31818 |
C00002526
|
||
| 5947 | RBP1, CRABP-I, CRBP, CRBP1, CRBPI, RBPC | retinol binding protein 1, cellular |
C00002526
|
| 5950 | RBP4, RDCCAS | retinol binding protein 4, plasma |
C00002526
|
| 5954 | RCN1, PIG20, RCAL, RCN | reticulocalbin 1, EF-hand calcium binding domain |
C00002526
|
| 7905 | REEP5, C5orf18, D5S346, DP1, TB2, YOP1 | receptor accessory protein 5 |
C00002526
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002526
|
| 6001 | RGS10 | regulator of G-protein signaling 10 |
C00002526
|
| 6004 | RGS16, A28-RGS14, A28-RGS14P, RGS-R | regulator of G-protein signaling 16 |
C00002526
|
| 9886 | RHOBTB1 | Rho-related BTB domain containing 1 |
C00002526
|
| 58480 | RHOU, ARHU, CDC42L1, DJ646B12.2, WRCH1, fJ646B12.2, hG28K | ras homolog family member U |
C00002526
|
| 116028 | RMI2, BLAP18, C16orf75 | RecQ mediated genome instability 2 |
C00002526
|
| 246243 | RNASEH1, H1RNA, RNH1 | ribonuclease H1 (EC:3.1.26.4) |
C00002526
|
| 8635 | RNASET2, RNASE6PL, bA514O12.3 | ribonuclease T2 (EC:3.1.27.1) |
C00002526
|
| 84900 | RNFT2, TMEM118 | ring finger protein, transmembrane 2 |
C00002526
|
| 55178 | RNMTL1 | RNA methyltransferase like 1 (EC:2.1.1.-) |
C00002526
|
| 6051 | RNPEP | arginyl aminopeptidase (aminopeptidase B) (EC:3.4.11.6) |
C00002526
|
| 83853 | ROPN1L, ASP, RSPH11 | rhophilin associated tail protein 1-like |
C00002526
|
| 130773 |
C00002526
|
||
| 6158 | RPL28, L28 | ribosomal protein L28 (EC:3.6.5.3) |
C00002526
|
| 6169 | RPL38, L38 | ribosomal protein L38 |
C00002526
|
| 6205 | RPS11, S11 | ribosomal protein S11 |
C00002526
|
| 6217 | RPS16, S16 | ribosomal protein S16 |
C00002526
|
| 6222 | RPS18, D6S218E, HKE3, KE-3, KE3, S18 | ribosomal protein S18 |
C00002526
|
| 6223 | RPS19, DBA, DBA1, S19 | ribosomal protein S19 |
C00002526
|
| 6230 | RPS25, S25 | ribosomal protein S25 |
C00002526
|
| 6235 | RPS29, S29 | ribosomal protein S29 |
C00002526
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00002526
|
| 6236 | RRAD, RAD, RAD1, REM3 | Ras-related associated with diabetes |
C00002526
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00002526
|
| 50484 | RRM2B, MTDPS8A, MTDPS8B, P53R2 | ribonucleotide reductase M2 B (TP53 inducible) (EC:1.17.4.1) |
C00002526
|
| 23223 | RRP12, KIAA0690 | ribosomal RNA processing 12 homolog (S. cerevisiae) |
C00002526
|
| 9136 | RRP9, RNU3IP2, U3-55K | ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
C00002526
|
| 23212 | RRS1 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
C00002526
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00002526
|
| 140576 | S100A16, DT1P1A7, S100F | S100 calcium binding protein A16 |
C00002526
|
| 6279 | S100A8, 60B8AG, CAGA, CFAG, CGLA, CP-10, L1Ag, MA387, MIF, MRP8, NIF, P8 | S100 calcium binding protein A8 |
C00002526
|
| 6280 | S100A9, 60B8AG, CAGB, CFAG, CGLB, L1AG, LIAG, MAC387, MIF, MRP14, NIF, P14 | S100 calcium binding protein A9 |
C00002526
|
| 1903 | S1PR3, EDG-3, EDG3, LPB3, S1P3 | sphingosine-1-phosphate receptor 3 |
C00002526
|
| 29901 | SAC3D1, HSU79266, SHD1 | SAC3 domain containing 1 |
C00002526
|
| 6299 | SALL1, HSAL1, Sal-1, TBS, ZNF794 | spalt-like transcription factor 1 |
C00002526
|
| 45384 |
C00002526
|
||
| 6309 | SC5D, ERG3, S5DES, SC5DL | sterol-C5-desaturase (EC:1.14.21.6) |
C00002526
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00002526
|
| 8796 | SCEL | sciellin |
C00002526
|
| 29970 | SCHIP1, SCHIP-1 | schwannomin interacting protein 1 |
C00002526
|
| 6334 | SCN8A, CERIII, CIAT, EIEE13, MED, NaCh6, Nav1.6, PN4 | sodium channel, voltage gated, type VIII, alpha subunit |
C00002526
|
| 6390 | SDHB, CWS2, IP, PGL4, SDH, SDH1, SDH2, SDHIP | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) (EC:1.3.5.1) |
C00002526
|
| 9871 | SEC24D | SEC24 family member D |
C00002526
|
| 29927 | SEC61A1, HSEC61, SEC61, SEC61A | Sec61 alpha 1 subunit (S. cerevisiae) |
C00002526
|
| 8991 | SELENBP1, LPSB, SBP56, SP56, hSBP | selenium binding protein 1 |
C00002526
|
| 22929 | SEPHS1, SELD, SPS, SPS1 | selenophosphate synthetase 1 (EC:2.7.9.3) |
C00002526
|
| 5265 | SERPINA1, A1A, A1AT, AAT, PI, PI1, PRO2275, alpha1AT | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
C00002526
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00002526
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00002526
|
| 83443 | SF3B5, SF3b10, Ysf3 | splicing factor 3b, subunit 5, 10kDa |
C00002526
|
| 6424 | SFRP4, FRP-4, FRPHE, sFRP-4 | secreted frizzled-related protein 4 |
C00002526
|
| 118980 | SFXN2 | sideroflexin 2 |
C00002526
|
| 6446 | SGK1, SGK | serum/glucocorticoid regulated kinase 1 (EC:2.7.11.1) |
C00002526
|
| 81537 | SGPP1, SPPase1 | sphingosine-1-phosphate phosphatase 1 |
C00002526
|
| 6462 | SHBG, ABP, SBP, TEBG | sex hormone-binding globulin |
C00002526
|
| 6474 | SHOX2, OG12, OG12X, SHOT | short stature homeobox 2 |
C00002526
|
| 6478 | SIAH2, hSiah2 | siah E3 ubiquitin protein ligase 2 (EC:6.3.2.-) |
C00002526
|
| 23411 | SIRT1, SIR2L1 | sirtuin 1 |
C00002526
|
| 6558 | SLC12A2, BSC, BSC2, NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
C00002526
|
| 6560 | SLC12A4, KCC1 | solute carrier family 12 (potassium/chloride transporter), member 4 |
C00002526
|
| 9123 | SLC16A3, MCT_3, MCT_4, MCT-3, MCT-4, MCT3, MCT4 | solute carrier family 16 (monocarboxylate transporter), member 3 |
C00002526
|
| 6573 | SLC19A1, CHMD, FOLT, IFC1, REFC, RFC1 | solute carrier family 19 (folate transporter), member 1 |
C00002526
|
| 6509 | SLC1A4, ASCT1, SATT | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
C00002526
|
| 6510 | SLC1A5, AAAT, ASCT2, ATBO, M7V1, M7VS1, R16, RDRC | solute carrier family 1 (neutral amino acid transporter), member 5 |
C00002526
|
| 51310 | SLC22A17, 24p3R, BOCT, BOIT, NGALR, NGALR2, NGALR3, hBOIT | solute carrier family 22, member 17 |
C00002526
|
| 6584 | SLC22A5, CDSP, OCTN2, OCTN2VT | solute carrier family 22 (organic cation/carnitine transporter), member 5 |
C00002526
|
| 9962 | SLC23A2, NBTL1, SLC23A1, SVCT2, YSPL2 | solute carrier family 23 (ascorbic acid transporter), member 2 |
C00002526
|
| 60386 | SLC25A19, DNC, MCPHA, MUP1, THMD3, THMD4, TPC | solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
C00002526
|
| 79751 | SLC25A22, EIEE3, GC1, NET44 | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
C00002526
|
| 115111 | SLC26A7, SUT2 | solute carrier family 26 (anion exchanger), member 7 |
C00002526
|
| 6513 | SLC2A1, DYT17, DYT18, DYT9, EIG12, GLUT, GLUT1, GLUT1DS, HTLVR, PED | solute carrier family 2 (facilitated glucose transporter), member 1 |
C00002526
|
| 7779 | SLC30A1, ZNT1, ZRC1 | solute carrier family 30 (zinc transporter), member 1 |
C00002526
|
| 1317 | SLC31A1, COPT1, CTR1 | solute carrier family 31 (copper transporter), member 1 |
C00002526
|
| 64116 | SLC39A8, BIGM103, LZT-Hs6, ZIP8 | solute carrier family 39 (zinc transporter), member 8 |
C00002526
|
| 6520 | SLC3A2, 4F2, 4F2HC, 4T2HC, CD98, CD98HC, MDU1, NACAE | solute carrier family 3 (amino acid transporter heavy chain), member 2 |
C00002526
|
| 146802 | SLC47A2, MATE2, MATE2-B, MATE2-K, MATE2K | solute carrier family 47 (multidrug and toxin extrusion), member 2 |
C00002526
|
| 6528 | SLC5A5, NIS, TDH1 | solute carrier family 5 (sodium/iodide cotransporter), member 5 |
C00002526
|
| 8884 | SLC5A6, SMVT | solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
C00002526
|
| 6539 | SLC6A12, BGT-1, BGT1, GAT2 | solute carrier family 6 (neurotransmitter transporter), member 12 |
C00002526
|
| 11254 | SLC6A14, BMIQ11 | solute carrier family 6 (amino acid transporter), member 14 |
C00002526
|
| 6533 | SLC6A6, TAUT | solute carrier family 6 (neurotransmitter transporter), member 6 |
C00002526
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00002526
|
| 8140 | SLC7A5, 4F2LC, CD98, D16S469E, E16, LAT1, MPE16, hLAT1 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
C00002526
|
| 81893 | SLC7A5P1, DC49, LAT1-3TM, MLAS, hLAT1-3TM | solute carrier family 7 (amino acid transporter light chain, L system), member 5 pseudogene 1 |
C00002526
|
| 387254 | SLC7A5P2, IMAA, MMAA | solute carrier family 7 (amino acid transporter light chain, L system), member 5 pseudogene 2 |
C00002526
|
| 9368 | SLC9A3R1, EBP50, NHERF, NHERF-1, NHERF1, NPHLOP2 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
C00002526
|
| 6578 | SLCO2A1, MATR1, OATP2A1, PGT, PHOAR2, SLC21A2 | solute carrier organic anion transporter family, member 2A1 |
C00002526
|
| 162394 | SLFN5 | schlafen family member 5 |
C00002526
|
| 6586 | SLIT3, MEGF5, SLIL2, SLIT1, Slit-3, slit2 | slit homolog 3 (Drosophila) |
C00002526
|
| 9748 | SLK, LOSK, STK2, bA16H23.1, se20-9 | STE20-like kinase (EC:2.7.11.1) |
C00002526
|
| 57228 | SMAGP, hSMAGP | small cell adhesion glycoprotein |
C00002526
|
| 6597 | SMARCA4, BAF190, BAF190A, BRG1, MRD16, RTPS2, SNF2, SNF2L4, SNF2LB, SWI2, hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
C00002526
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00002526
|
| 10051 | SMC4, CAP-C, CAPC, SMC-4, SMC4L1, hCAP-C | structural maintenance of chromosomes 4 |
C00002526
|
| 23137 | SMC5, SMC5L1 | structural maintenance of chromosomes 5 |
C00002526
|
| 23049 | SMG1, 61E3.4, ATX, LIP | SMG1 phosphatidylinositol 3-kinase-related kinase (EC:2.7.11.1) |
C00002526
|
| 201895 | SMIM14, C4orf34 | small integral membrane protein 14 |
C00002526
|
| 54498 | SMOX, C20orf16, PAO, PAO-1, PAOH1, SMO | spermine oxidase (EC:1.5.3.16) |
C00002526
|
| 724102 | SNHG4, NCRNA00059, U19H | small nucleolar RNA host gene 4 (non-protein coding) |
C00002526
|
| 677838 | SNORA61, ACA61 | small nucleolar RNA, H/ACA box 61 |
C00002526
|
| 6043 | SNORA63, E3, E3-2, RNE3, RNU107 | small nucleolar RNA, H/ACA box 63 |
C00002526
|
| 6626 | SNRPA, Mud1, U1-A, U1A | small nuclear ribonucleoprotein polypeptide A |
C00002526
|
| 9021 | SOCS3, ATOD4, CIS3, Cish3, SOCS-3, SSI-3, SSI3 | suppressor of cytokine signaling 3 |
C00002526
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00002526
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002526
|
| 6649 | SOD3, EC-SOD | superoxide dismutase 3, extracellular (EC:1.15.1.1) |
C00002526
|
| 10580 | SORBS1, CAP, FLAF2, R85FL, SH3D5, SH3P12, SORB1 | sorbin and SH3 domain containing 1 |
C00002526
|
| 6652 | SORD, SORD1 | sorbitol dehydrogenase (EC:1.1.1.14) |
C00002526
|
| 6657 | SOX2, ANOP3, MCOPS3 | SRY (sex determining region Y)-box 2 |
C00002526
|
| 6659 | SOX4, EVI16 | SRY (sex determining region Y)-box 4 |
C00002526
|
| 6667 | SP1 | Sp1 transcription factor |
C00002526
|
| 79582 | SPAG16, PF20, WDR29 | sperm associated antigen 16 |
C00002526
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00002526
|
| 124044 | SPATA2L, C16orf76, tamo | spermatogenesis associated 2-like |
C00002526
|
| 25803 | SPDEF, PDEF, RP11-375E1__A.3, bA375E1.3 | SAM pointed domain containing ETS transcription factor |
C00002526
|
| 10927 | SPIN1, SPIN | spindlin 1 |
C00002526
|
| 90864 | SPSB3, C16orf31, SSB3 | splA/ryanodine receptor domain and SOCS box containing 3 |
C00002526
|
| 6713 | SQLE | squalene epoxidase (EC:1.14.13.132) |
C00002526
|
| 8878 | SQSTM1, A170, OSIL, PDB3, ZIP3, p60, p62, p62B | sequestosome 1 |
C00002526
|
| 136853 | SRCRB4D, S4D-SRCRB, SRCRB-S4D, SSC4D | scavenger receptor cysteine rich domain containing, group B (4 domains) |
C00002526
|
| 6716 | SRD5A2 | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) (EC:1.3.1.22) |
C00002526
|
| 79644 | SRD5A3, CDG1P, CDG1Q, KRIZI, SRD5A2L, SRD5A2L1 | steroid 5 alpha-reductase 3 (EC:1.3.1.22 1.3.1.94) |
C00002526
|
| 6723 | SRM, PAPT, SPDSY, SPS1, SRML1 | spermidine synthase (EC:2.5.1.16) |
C00002526
|
| 6734 | SRPR, DP, Sralpha | signal recognition particle receptor (docking protein) |
C00002526
|
| 140809 | SRXN1, C20orf139, Npn3, SRX, SRX1 | sulfiredoxin 1 (EC:1.8.98.2) |
C00002526
|
| 6770 | STAR, STARD1 | steroidogenic acute regulatory protein |
C00002526
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00002526
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00002526
|
| 6781 | STC1, STC | stanniocalcin 1 |
C00002526
|
| 8614 | STC2, STC-2, STCRP | stanniocalcin 2 |
C00002526
|
| 26872 | STEAP1, PRSS24, STEAP | six transmembrane epithelial antigen of the prostate 1 |
C00002526
|
| 55240 | STEAP3, AHMIO2, STMP3, TSAP6, dudlin-2 | STEAP family member 3, metalloreductase |
C00002526
|
| 27347 | STK39, DCHT, PASK, SPAK | serine threonine kinase 39 (EC:2.7.11.1) |
C00002526
|
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00002526
|
| 6812 | STXBP1, MUNC18-1, NSEC1, P67, RBSEC1, UNC18 | syntaxin binding protein 1 |
C00002526
|
| 55959 | SULF2, HSULF-2 | sulfatase 2 |
C00002526
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00002526
|
| 6818 | SULT1A3, HAST, HAST3, M-PST, ST1A3/ST1A4, ST1A5, STM, TL-PST | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (EC:2.8.2.1) |
C00002526
|
| 6783 | SULT1E1, EST, EST-1, ST1E1, STE | sulfotransferase family 1E, estrogen-preferring, member 1 (EC:2.8.2.4) |
C00002526
|
| 6836 | SURF4, ERV29 | surfeit 4 |
C00002526
|
| 203328 | SUSD3 | sushi domain containing 3 |
C00002526
|
| 94056 | SYAP1 | synapse associated protein 1 |
C00002526
|
| 8871 | SYNJ2, INPP5H | synaptojanin 2 (EC:3.1.3.36) |
C00002526
|
| 10460 | TACC3, ERIC-1, ERIC1 | transforming, acidic coiled-coil containing protein 3 |
C00002526
|
| 6870 | TACR3, HH11, NK-3R, NK3R, NKR, TAC3RL | tachykinin receptor 3 |
C00002526
|
| 4070 | TACSTD2, EGP-1, EGP1, GA733-1, GA7331, GP50, M1S1, TROP2 | tumor-associated calcium signal transducer 2 |
C00002526
|
| 22980 | TCF25, Hulp1, NULP1, PRO2620, hKIAA1049 | transcription factor 25 (basic helix-loop-helix) |
C00002526
|
| 6925 | TCF4, E2-2, ITF-2, ITF2, PTHS, SEF-2, SEF2, SEF2-1, SEF2-1A, SEF2-1B, TCF-4, bHLHb19 | transcription factor 4 |
C00002526
|
| 7005 | TEAD3, DTEF-1, ETFR-1, TEAD-3, TEAD5, TEF-5, TEF5 | TEA domain family member 3 |
C00002526
|
| 9894 | TELO2, CLK2, TEL2 | telomere maintenance 2 |
C00002526
|
| 7015 | TERT, CMM9, DKCA2, DKCB4, EST2, PFBMFT1, TCS1, TP2, TRT, hEST2, hTRT | telomerase reverse transcriptase (EC:2.7.7.49) |
C00002526
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00002526
|
| 7033 | TFF3, ITF, P1B, TFI | trefoil factor 3 (intestinal) |
C00002526
|
| 7035 | TFPI, EPI, LACI, TFI, TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
C00002526
|
| 7037 | TFRC, CD71, T9, TFR, TFR1, TR, TRFR, p90 | transferrin receptor |
C00002526
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002526
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00002526
|
| 7042 | TGFB2, LDS4, TGF-beta2 | transforming growth factor, beta 2 |
C00002526
|
| 7043 | TGFB3, ARVD, ARVD1, TGF-beta3 | transforming growth factor, beta 3 |
C00002526
|
| 7057 | THBS1, THBS, THBS-1, TSP, TSP-1, TSP1 | thrombospondin 1 |
C00002526
|
| 51337 | THEM6, C8orf55, DSCD75 | thioesterase superfamily member 6 |
C00002526
|
| 57187 | THOC2, CXorf3, THO2, dJ506G2.1, hTREX120 | THO complex 2 |
C00002526
|
| 7072 | TIA1, TIA-1, WDM | TIA1 cytotoxic granule-associated RNA binding protein |
C00002526
|
| 9220 | TIAF1, MAJN, SPR210 | TGFB1-induced anti-apoptotic factor 1 |
C00002526
|
| 26515 | TIMM10B, FXC1, TIM10B, Tim9b | translocase of inner mitochondrial membrane 10 homolog B (yeast) |
C00002526
|
| 7076 | TIMP1, CLGI, EPA, EPO, HCI, TIMP | TIMP metallopeptidase inhibitor 1 |
C00002526
|
| 7077 | TIMP2, CSC-21K, DDC8 | TIMP metallopeptidase inhibitor 2 |
C00002526
|
| 7078 | TIMP3, HSMRK222, K222, K222TA2, SFD | TIMP metallopeptidase inhibitor 3 |
C00002526
|
| 26277 | TINF2, DKCA3, TIN2 | TERF1 (TRF1)-interacting nuclear factor 2 |
C00002526
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00002526
|
| 7086 | TKT, TK, TKT1 | transketolase (EC:2.2.1.1) |
C00002526
|
| 7093 | TLL2 | tolloid-like 2 |
C00002526
|
| 9911 | TMCC2, HUCEP11 | transmembrane and coiled-coil domain family 2 |
C00002526
|
| 54732 | TMED9, GMP25, HSGP25L2G, p25 | transmembrane emp24 protein transport domain containing 9 |
C00002526
|
| 83862 | TMEM120A, NET29, TMPIT | transmembrane protein 120A |
C00002526
|
| 80757 | TMEM121, hole | transmembrane protein 121 |
C00002526
|
| 135932 | TMEM139 | transmembrane protein 139 |
C00002526
|
| 55151 | TMEM38B, C9orf87, D4Ertd89e, OI14, TRIC-B, TRICB, bA219P18.1 | transmembrane protein 38B |
C00002526
|
| 55287 | TMEM40 | transmembrane protein 40 |
C00002526
|
| 120224 | TMEM45B | transmembrane protein 45B |
C00002526
|
| 113452 | TMEM54, BCLP, CAC-1, CAC1 | transmembrane protein 54 |
C00002526
|
| 27346 | TMEM97, MAC30 | transmembrane protein 97 |
C00002526
|
| 7112 | TMPO, CMD1T, LAP2, LEMD4, PRO0868, TP | thymopoietin |
C00002526
|
| 7113 | TMPRSS2, PP9284, PRSS10 | transmembrane protease, serine 2 (EC:3.4.21.-) |
C00002526
|
| 83857 | TMTC1, OLF, TMTC1A | transmembrane and tetratricopeptide repeat containing 1 |
C00002526
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002526
|
| 7130 | TNFAIP6, TSG-6, TSG6 | tumor necrosis factor, alpha-induced protein 6 |
C00002526
|
| 25816 | TNFAIP8, GG2-1, MDC-3.13, NDED, SCC-S2, SCCS2 | tumor necrosis factor, alpha-induced protein 8 |
C00002526
|
| 8795 | TNFRSF10B, CD262, DR5, KILLER, KILLER/DR5, TRAIL-R2, TRAILR2, TRICK2, TRICK2A, TRICK2B, TRICKB, ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b |
C00002526
|
| 4982 | TNFRSF11B, OCIF, OPG, TR1 | tumor necrosis factor receptor superfamily, member 11b |
C00002526
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00002526
|
| 7132 | TNFRSF1A, CD120a, FPF, MS5, TBP1, TNF-R, TNF-R-I, TNF-R55, TNFAR, TNFR1, TNFR1-d2, TNFR55, TNFR60, p55, p55-R, p60 | tumor necrosis factor receptor superfamily, member 1A |
C00002526
|
| 8718 | TNFRSF25, APO-3, DDR3, DR3, LARD, TNFRSF12, TR3, TRAMP, WSL-1, WSL-LR | tumor necrosis factor receptor superfamily, member 25 |
C00002526
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00002526
|
| 7292 | TNFSF4, CD134L, CD252, GP34, OX-40L, OX4OL, TXGP1 | tumor necrosis factor (ligand) superfamily, member 4 |
C00002526
|
| 8744 | TNFSF9, 4-1BB-L, CD137L | tumor necrosis factor (ligand) superfamily, member 9 |
C00002526
|
| 85456 | TNKS1BP1, TAB182 | tankyrase 1 binding protein 1, 182kDa |
C00002526
|
| 7138 | TNNT1, ANM, NEM5, STNT, TNT, TNTS | troponin T type 1 (skeletal, slow) |
C00002526
|
| 10452 | TOMM40, C19orf1, D19S1177E, PER-EC1, PEREC1, TOM40 | translocase of outer mitochondrial membrane 40 homolog (yeast) |
C00002526
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00002526
|
| 7155 | TOP2B, TOPIIB, top2beta | topoisomerase (DNA) II beta 180kDa (EC:5.99.1.3) |
C00002526
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00002526
|
| 9537 | TP53I11, PIG11 | tumor protein p53 inducible protein 11 |
C00002526
|
| 9540 | TP53I3, PIG3 | tumor protein p53 inducible protein 3 |
C00002526
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00002526
|
| 8626 | TP63, AIS, B(p51A), B(p51B), EEC3, KET, LMS, NBP, OFC8, RHS, SHFM4, TP53CP, TP53L, TP73L, p40, p51, p53CP, p63, p73H, p73L | tumor protein p63 |
C00002526
|
| 7164 | TPD52L1, D53, hD53 | tumor protein D52-like 1 |
C00002526
|
| 27010 | TPK1, HTPK1, PP20, THMD5 | thiamin pyrophosphokinase 1 (EC:2.7.6.2) |
C00002526
|
| 7168 | TPM1, C15orf13, CMD1Y, CMH3, HTM-alpha, LVNC9, TMSA | tropomyosin 1 (alpha) |
C00002526
|
| 7170 | TPM3, CAPM1, CFTD, NEM1, OK/SW-cl.5, TM-5, TM3, TM30, TM30nm, TM5, TPMsk3, TRK, hscp30 | tropomyosin 3 |
C00002526
|
| 7173 | TPO, MSA, TDH2A, TPX | thyroid peroxidase (EC:1.11.1.8) |
C00002526
|
| 7175 | TPR | translocated promoter region, nuclear basket protein |
C00002526
|
| 10626 | TRIM16, EBBP | tripartite motif containing 16 |
C00002526
|
| 7706 | TRIM25, EFP, RNF147, Z147, ZNF147 | tripartite motif containing 25 (EC:6.3.2.19 6.3.2.n3) |
C00002526
|
| 129868 | TRIM43, TRIM43A | tripartite motif containing 43 |
C00002526
|
| 653192 | TRIM43B | tripartite motif containing 43B |
C00002526
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00002526
|
| 51504 | TRMT112, HSPC152, HSPC170, TRM112, TRMT11-2 | tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
C00002526
|
| 8848 | TSC22D1, Ptg-2, TGFB1I4, TSC22 | TSC22 domain family, member 1 |
C00002526
|
| 25987 | TSKU, E2IG4, LRRC54, TSK | tsukushi, small leucine rich proteoglycan |
C00002526
|
| 23554 | TSPAN12, EVR5, NET-2, NET2, TM4SF12 | tetraspanin 12 |
C00002526
|
| 146057 | TTBK2, SCA11, TTBK | tau tubulin kinase 2 (EC:2.7.11.1) |
C00002526
|
| 283237 | TTC9C | tetratricopeptide repeat domain 9C |
C00002526
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00002526
|
| 23170 | TTLL12, dJ526I14.2 | tubulin tyrosine ligase-like family, member 12 |
C00002526
|
| 7277 | TUBA4A, H2-ALPHA, TUBA1 | tubulin, alpha 4a |
C00002526
|
| 203068 | TUBB, M40, OK/SW-cl.56, TUBB1, TUBB5 | tubulin, beta class I |
C00002526
|
| 84617 | TUBB6, HsT1601, TUBB-5 | tubulin, beta 6 class V |
C00002526
|
| 7289 | TULP3, TUBL3 | tubby like protein 3 |
C00002526
|
| 200081 | TXLNA, IL14, RP4-622L5.4, TXLN | taxilin alpha |
C00002526
|
| 7295 | TXN, TRDX, TRX, TRX1 | thioredoxin |
C00002526
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00002526
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00002526
|
| 7306 | TYRP1, CAS2, CATB, GP75, OCA3, TRP, TRP1, TYRP, b-PROTEIN | tyrosinase-related protein 1 (EC:1.14.18.-) |
C00002526
|
| 6675 | UAP1, AGX, AGX1, AGX2, SPAG2 | UDP-N-acetylglucosamine pyrophosphorylase 1 (EC:2.7.7.23 2.7.7.83) |
C00002526
|
| 9898 | UBAP2L, NICE-4 | ubiquitin associated protein 2-like |
C00002526
|
| 11065 | UBE2C, UBCH10, dJ447F3.2 | ubiquitin-conjugating enzyme E2C (EC:6.3.2.19) |
C00002526
|
| 9246 | UBE2L6, RIG-B, UBCH8 | ubiquitin-conjugating enzyme E2L 6 (EC:6.3.2.19) |
C00002526
|
| 29089 | UBE2T, PIG50 | ubiquitin-conjugating enzyme E2T (putative) (EC:6.3.2.19) |
C00002526
|
| 56893 | UBQLN4, A1U, A1Up, C1orf6, CIP75, UBIN | ubiquilin 4 |
C00002526
|
| 7343 | UBTF, NOR-90, UBF, UBF-1, UBF1 | upstream binding transcription factor, RNA polymerase I |
C00002526
|
| 7345 | UCHL1, NDGOA, PARK5, PGP_9.5, PGP9.5, PGP95, Uch-L1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (EC:3.4.19.12) |
C00002526
|
| 7358 | UGDH, GDH, UDP-GlcDH, UDPGDH, UGD | UDP-glucose 6-dehydrogenase (EC:1.1.1.22) |
C00002526
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00002526
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00002526
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00002526
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00002526
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00002526
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00002526
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00002526
|
| 7367 | UGT2B17, BMND12, UDPGT2B17 | UDP glucuronosyltransferase 2 family, polypeptide B17 (EC:2.4.1.17) |
C00002526
|
| 29128 | UHRF1, ICBP90, Np95, RNF106, hNP95, hUHRF1 | ubiquitin-like with PHD and ring finger domains 1 (EC:6.3.2.19) |
C00002526
|
| 7374 | UNG, DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 | uracil-DNA glycosylase (EC:3.2.2.27) |
C00002526
|
| 10868 | USP20, LSFR3A, VDU2, hVDU2 | ubiquitin specific peptidase 20 (EC:3.4.19.12) |
C00002526
|
| 114990 | VASN, SLITL2 | vasorin |
C00002526
|
| 10493 | VAT1, VATI | vesicle amine transport 1 |
C00002526
|
| 7409 | VAV1, VAV | vav 1 guanine nucleotide exchange factor |
C00002526
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002526
|
| 7414 | VCL, CMD1W, CMH15, MV, MVCL | vinculin |
C00002526
|
| 79609 | VCPKMT, C14orf138, METTL21D, VCP-KMT | valosin containing protein lysine (K) methyltransferase |
C00002526
|
| 7421 | VDR, NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
C00002526
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00002526
|
| 40542 |
C00002526
|
||
| 7453 | WARS, GAMMA-2, IFI53, IFP53 | tryptophanyl-tRNA synthetase (EC:6.1.1.2) |
C00002526
|
| 10885 | WDR3, DIP2, UTP12 | WD repeat domain 3 |
C00002526
|
| 10785 | WDR4, TRM82, TRMT82 | WD repeat domain 4 |
C00002526
|
| 8839 | WISP2, CCN5, CT58, CTGF-L | WNT1 inducible signaling pathway protein 2 |
C00002526
|
| 7476 | WNT7A | wingless-type MMTV integration site family, member 7A |
C00002526
|
| 331 | XIAP, API3, BIRC4, IAP-3, ILP1, MIHA, XLP2, hIAP-3, hIAP3 | X-linked inhibitor of apoptosis |
C00002526
|
| 51646 | YPEL5 | yippee-like 5 (Drosophila) |
C00002526
|
| 79413 | ZBED2 | zinc finger, BED-type containing 2 |
C00002526
|
| 84872 | ZC3H10, ZC3HDC10 | zinc finger CCCH-type containing 10 |
C00002526
|
| 677 | ZFP36L1, BRF1, Berg36, ERF-1, ERF1, RNF162B, TIS11B, cMG1 | ZFP36 ring finger protein-like 1 |
C00002526
|
| 23414 | ZFPM2, DIH3, FOG2, ZC2HC11B, ZNF89B, hFOG-2 | zinc finger protein, FOG family member 2 |
C00002526
|
| 342357 | ZKSCAN2, ZNF694, ZSCAN31, ZSCAN34 | zinc finger with KRAB and SCAN domains 2 |
C00002526
|
| 9203 | ZMYM3, DXS6673E, MYM, XFIM, ZNF198L2, ZNF261 | zinc finger, MYM-type 3 |
C00002526
|
| 7730 | ZNF177, PIGX | zinc finger protein 177 |
C00002526
|
| 7587 | ZNF37A, KOX21, ZNF37 | zinc finger protein 37A |
C00002526
|
| 80139 | ZNF703, ZEPPO1, ZNF503L, ZPO1 | zinc finger protein 703 |
C00002526
|
| 11130 | ZWINT, HZwint-1, KNTC2AP, ZWINT1 | ZW10 interacting kinetochore protein |
C00002526
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (117)
| OMIM | preferred title | UniProt |
|---|---|---|
| #100100 | Abdominal muscles, absence of, with urinary tract abnormality and cryptorchidism |
P20309
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #103780 | Alcohol dependence |
P08172
P14416 P31645 |
| #610251 | Alcohol sensitivity, acute |
P05091
|
| #614373 | Amyotrophic lateral sclerosis 16, juvenile; als16 |
Q99720
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #602025 | Body mass index quantitative trait locus 9; bmiq9 |
P41968
|
| %606641 | Body mass index; bmi |
P37231
|
| #114480 | Breast cancer |
P31749
P38398 |
| #604370 | Breast-ovarian cancer, familial, susceptibility to, 1; brovca1 |
P38398
|
| #300615 | Brunner syndrome |
P21397
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #114500 | Colorectal cancer; crc |
P31749
|
| #615109 | Cowden syndrome 6; cws6 |
P31749
|
| #162800 | Cyclic neutropenia |
P08246
|
| #608565 | Deafness, autosomal recessive 35; dfnb35 |
O95718
|
| #612522 | Diabetes mellitus, insulin-dependent, 22; iddm22 |
P51681
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #133239 | Esophageal cancer |
P04637
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613659 | Gastric cancer |
P04626
|
| #137215 | Gastric cancer, hereditary diffuse; hdgc |
P04626
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #231095 | Ghosal hematodiaphyseal dysplasia; ghdd |
P24557
|
| #137800 | Glioma susceptibility 1; glm1 |
P04626
P37231 |
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #609423 | Human immunodeficiency virus type 1, susceptibility to |
P41597
P51681 |
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237450 | Hyperbilirubinemia, rotor type; hblrr |
Q9NPD5
Q9Y6L6 |
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #608232 | Leukemia, chronic myeloid; cml |
P00519
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #211980 | Lung cancer |
P00533
P04626 P04637 |
| #608516 | Major depressive disorder; mdd |
P08172
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #609048 | Melanoma, cutaneous malignant, susceptibility to, 3; cmm3 |
P11802
|
| %300852 | Mental retardation, x-linked 88; mrx88 |
P50052
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #126200 | Multiple sclerosis, susceptibility to; ms |
P08575
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #254500 | Myeloma, multiple |
P24385
|
| #159900 | Myoclonic dystonia |
P14416
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #202700 | Neutropenia, severe congenital, 1, autosomal dominant; scn1 |
P08246
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #257220 | Niemann-pick disease, type c1; npc1 |
O15118
|
| #601665 | Obesity |
P32245
P37231 |
| #164230 | Obsessive-compulsive disorder; ocd |
P31645
|
| #604715 | Orthostatic intolerance |
P23975
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #167000 | Ovarian cancer |
P04626
P38398 |
| #614320 | Pancreatic cancer, susceptibility to, 4; pnca4 |
P38398
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #613135 | Parkinsonism-dystonia, infantile; pkdys |
Q01959
|
| #172700 | Pick disease of brain |
P10636
|
| #176920 | Proteus syndrome |
P31749
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #608971 | Severe combined immunodeficiency, autosomal recessive, t cell-negative, b cell-positive, nk cell-positive |
P08575
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #187950 | Thrombocythemia 1; thcyt1 |
P40225
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #190300 | Tremor, hereditary essential, 1; etm1 |
P35462
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #610379 | West nile virus, susceptibility to |
P51681
|
| #278300 | Xanthinuria, type i |
P47989
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (106)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00136 | Niemann-Pick disease type C (NPC) |
O15118
(related)
|
| H00605 | Deafness, autosomal recessive |
O95718
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
P00519
(related)
P00519 (marker) Q03164 (related) Q03164 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
P00519
(related)
P00519 (marker) P04637 (related) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) P24385 (related) P24385 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P04637 (related) P04637 (marker) P24385 (related) P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04626 (related) P04637 (related) |
| H00022 | Bladder cancer |
P00533
(related)
P04626 (related) P04637 (related) |
| H00028 | Choriocarcinoma |
P00533
(related)
P03956 (related) P04626 (related) P04637 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P04626 (related) P11802 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) P11802 (related) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) P24385 (related) P24385 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04626 (related) P04637 (related) Q92731 (marker) |
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
P11511 (related) |
| H00019 | Pancreatic cancer |
P04626
(related)
P04637 (related) P04637 (marker) |
| H00027 | Ovarian cancer |
P04626
(related)
P04637 (related) P38398 (related) |
| H00031 | Breast cancer |
P04626
(related)
P04626 (marker) P04637 (related) P24385 (related) P38398 (related) |
| H00046 | Cholangiocarcinoma |
P04626
(related)
P04637 (related) P35354 (related) |
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
|
| H00006 | Hairy-cell leukemia |
P04637
(related)
P24385 (related) |
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
P24385 (related) |
| H00013 | Small cell lung cancer |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) |
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P14780 (related) P35354 (related) |
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
P37231 (related) |
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
P11802 (related) |
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
|
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00047 | Gallbladder cancer |
P04637
(related)
P24385 (marker) |
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00021 | Renal cell carcinoma |
P04637
(marker)
|
| H01071 | Acute alcohol sensitivity |
P05091
(related)
|
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H00100 | Neutropenic disorders |
P08246
(related)
|
| H00091 | T-B+Severe combined immunodeficiencies (SCIDs) |
P08575
(related)
|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H01031 | Orthostatic intolerance (OI) |
P23975
(related)
|
| H00559 | von Hippel-Lindau syndrome |
P24385
(related)
|
| H00490 | Diaphyseal dysplasia with anemia (Ghosal) |
P24557
(related)
|
| H00539 | PTEN hamartoma tumor syndrome (PHTS) |
P31749
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00227 | Congenital amegakaryocytic thrombocytopenia (CAMT) |
P40225
(marker)
|
| H00192 | Xanthinuria |
P47989
(related)
Q06278 (related) |
| H00480 | Non-syndromic X-linked mental retardation |
P50052
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00208 | Hyperbilirubinemia |
Q92887
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
Q9NUW8 (related) |
Diseases related to CTD interactions
38 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000014 | Abnormalities, Drug-Induced |
C00002526
|
| D000022 | Abortion, Spontaneous |
C00002526
|
| D000230 | Adenocarcinoma |
C00002526
|
| C538231 | Adenocarcinoma of lung |
C00002526
|
| D001836 | Body Weight Changes |
C00002526
|
| D001943 | Breast Neoplasms |
C00002526
|
| D006528 | Carcinoma, Hepatocellular |
C00002526
|
| D002318 | Cardiovascular Diseases |
C00002526
|
| D003110 | Colonic Neoplasms |
C00002526
|
| D000013 | Congenital Abnormalities |
C00002526
|
| D003924 | Diabetes Mellitus, Type 2 |
C00002526
|
| D012734 | Disorders of Sex Development |
C00002526
|
| D004487 | Edema |
C00002526
|
| D004714 | Endometrial Hyperplasia |
C00002526
|
| D016889 | Endometrial Neoplasms |
C00002526
|
| D005909 | Glioblastoma |
C00002526
|
| D006402 | Hematologic Diseases |
C00002526
|
| D006965 | Hyperplasia |
C00002526
|
| D007938 | Leukemia |
C00002526
|
| D017254 | Leukemic Infiltration |
C00002526
|
| D008104 | Liver Cirrhosis, Alcoholic |
C00002526
|
| D008106 | Liver Cirrhosis, Experimental |
C00002526
|
| D008325 | Mammary Neoplasms, Experimental |
C00002526
|
| D008679 | Metaplasia |
C00002526
|
| D009203 | Myocardial Infarction |
C00002526
|
| D015428 | Myocardial Reperfusion Injury |
C00002526
|
| D009336 | Necrosis |
C00002526
|
| D009369 | Neoplasms |
C00002526
|
| D009421 | Nervous System Malformations |
C00002526
|
| D009845 | Oligospermia |
C00002526
|
| D010024 | Osteoporosis |
C00002526
|
| D015663 | OSTEOPOROSIS, POSTMENOPAUSAL |
C00002526
|
| D010051 | Ovarian Neoplasms |
C00002526
|
| D047928 | Premature Birth |
C00002526
|
| D011297 | Prenatal Exposure Delayed Effects |
C00002526
|
| D011471 | Prostatic Neoplasms |
C00002526
|
| D013959 | Thyroid Diseases |
C00002526
|
| D014594 | Uterine Neoplasms |
C00002526
|