| Entry |
|
|---|
| Name |
L-Glutamate;
L-Glutamic acid;
L-Glutaminic acid;
Glutamate
|
|---|
| Formula |
C5H9NO4
|
|---|
| Exact mass |
147.0532
|
|---|
| Mol weight |
147.1293
|
|---|
| Structure |
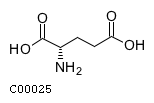
|
|---|
| Remark |
|
|---|
| Reaction |
|
|---|
| Pathway |
| map00250 | Alanine, aspartate and glutamate metabolism |
| map00330 | Arginine and proline metabolism |
| map00430 | Taurine and hypotaurine metabolism |
| map00524 | Neomycin, kanamycin and gentamicin biosynthesis |
| map00630 | Glyoxylate and dicarboxylate metabolism |
| map00660 | C5-Branched dibasic acid metabolism |
| map00997 | Biosynthesis of various other secondary metabolites |
| map00998 | Biosynthesis of various antibiotics |
| map01060 | Biosynthesis of plant secondary metabolites |
| map01064 | Biosynthesis of alkaloids derived from ornithine, lysine and nicotinic acid |
| map01110 | Biosynthesis of secondary metabolites |
| map01120 | Microbial metabolism in diverse environments |
| map01210 | 2-Oxocarboxylic acid metabolism |
| map04072 | Phospholipase D signaling pathway |
| map04080 | Neuroactive ligand-receptor interaction |
| map04723 | Retrograde endocannabinoid signaling |
| map04964 | Proximal tubule bicarbonate reclamation |
| map04974 | Protein digestion and absorption |
| map05022 | Pathways of neurodegeneration - multiple diseases |
| map05230 | Central carbon metabolism in cancer |
|
|---|
| Module |
| M00015 | Proline biosynthesis, glutamate => proline |
| M00027 | GABA (gamma-Aminobutyrate) shunt |
| M00028 | Ornithine biosynthesis, glutamate => ornithine |
| M00045 | Histidine degradation, histidine => N-formiminoglutamate => glutamate |
| M00118 | Glutathione biosynthesis, glutamate => glutathione |
| M00121 | Heme biosynthesis, plants and bacteria, glutamate => heme |
| M00763 | Ornithine biosynthesis, mediated by LysW, glutamate => ornithine |
| M00845 | Arginine biosynthesis, glutamate => acetylcitrulline => arginine |
| M00879 | Arginine succinyltransferase pathway, arginine => glutamate |
| M00970 | Proline degradation, proline => glutamate |
|
|---|
| Network |
|
|---|
| Enzyme |
|
|---|
| Brite |
Compounds with biological roles [BR:br08001]
Peptides
Amino acids
Common amino acids
C00025 Glutamic acid (Glu)
Hormones and transmitters
Neurotransmitters
Amino acids
C00025 Glutamate
Risk category of Japanese OTC drugs [BR:br08312]
Third-class OTC drugs
Inorganic and organic chemicals
Glutamic acid
D00007 Glutamic acid (USP)
Drugs listed in the Japanese Pharmacopoeia [BR:br08311]
Chemicals
D00007 L-Glutamic acid
|
|---|
| Other DBs |
|
|---|
| KCF data |
ATOM 10
1 C1c C 23.8372 -17.4608
2 C1b C 25.0252 -16.7233
3 C6a C 22.6023 -16.7994
4 N1a N 23.8781 -18.8595
5 C1b C 26.2601 -17.3788
6 O6a O 21.4434 -17.5954
7 O6a O 22.6198 -15.4007
8 C6a C 27.4482 -16.6414
9 O6a O 28.6830 -17.3028
10 O6a O 27.4714 -15.2426
BOND 9
1 1 2 1
2 1 3 1
3 1 4 1 #Down
4 2 5 1
5 3 6 1
6 3 7 2
7 5 8 1
8 8 9 1
9 8 10 2
|
|---|

