| Entry |
|
|---|
| Name |
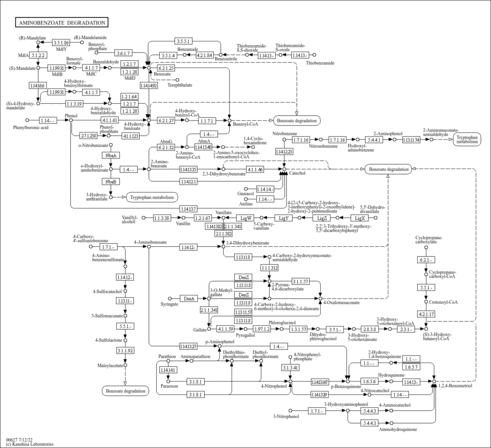
Aminobenzoate degradation |
|---|
| Class |
Metabolism; Xenobiotics biodegradation and metabolism
|
|---|
| Pathway map |
|
|---|
| Module |
| M00637 | Anthranilate degradation, anthranilate => catechol [PATH:map00627] |
|
|---|
| Other DBs |
|
|---|
| Reference |
|
|---|
| Authors |
Fewson CA |
|---|
| Title |
Microbial metabolism of mandelate: a microcosm of diversity. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
McLeish MJ, Kneen MM, Gopalakrishna KN, Koo CW, Babbitt PC, Gerlt JA, Kenyon GL |
|---|
| Title |
Identification and characterization of a mandelamide hydrolase and an NAD(P)+-dependent benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Shimao M, Nakamura T, Okuda A, Harayama S. |
|---|
| Title |
Characteristics of transposon insertion mutants of mandelic acid-utilizing Pseudomonas putida strain A10L. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Naidu D, Ragsdale SW. |
|---|
| Title |
Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Toraya T, Oka T, Ando M, Yamanishi M, Nishihara H. |
|---|
| Title |
Novel pathway for utilization of cyclopropanecarboxylate by Rhodococcus rhodochrous. |
|---|
| Journal |
|
|---|
Related
pathway |
| map00361 | Chlorocyclohexane and chlorobenzene degradation |
|
|---|
| KO pathway |
|
|---|