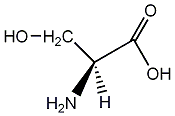
Metabolite
KNApSAcK Entry
| id | C00001393 |
|---|---|
| Name | L-Serine |
| CAS RN | 56-45-1 |
| Standard InChI | InChI=1S/C3H7NO3/c4-2(1-5)3(6)7/h2,5H,1,4H2,(H,6,7)/t2-/m0/s1 |
| Standard InChI (Main Layer) | InChI=1S/C3H7NO3/c4-2(1-5)3(6)7/h2,5H,1,4H2,(H,6,7) |
Cluster
| Phytochemical cluster | |
|---|---|
| KCF-S cluster | No. 3420 |
Link
ChEMBL
| By standard InChI | CHEMBL11298 |
|---|---|
| By standard InChI Main Layer | CHEMBL11298 CHEMBL285123 |
KEGG
| By LinkDB | C00065 |
|---|
CTD
| By CAS RN |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 1 |
Family
| family name | count |
|---|---|
| Brassicaceae | 1 |
| Enterobacteriaceae | 1 |
List (2)
* NCBI
| KNApSAcK organism | *ID | *family | *plant class | *kingdom |
|---|---|---|---|---|
| Arabidopsis thaliana | 3702 | Brassicaceae | rosids | Viridiplantae |
| Escherichia coli | 562 | Enterobacteriaceae | Bacteria |
Human Protein / Gene in interaction
13 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | CHEMBL11298 |
CHEMBL1741321
(3)
|
1 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | CHEMBL285123 |
CHEMBL1794499
(1)
|
2 / 0 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | CHEMBL285123 |
CHEMBL1794573
(1)
|
2 / 2 |
| P02545 | Prelamin-A/C | Unclassified protein | CHEMBL285123 |
CHEMBL1614544
(1)
|
11 / 10 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | CHEMBL11298 |
CHEMBL1741325
(3)
|
0 / 1 |
| P54132 | Bloom syndrome protein | Enzyme | CHEMBL285123 |
CHEMBL1614522
(1)
CHEMBL1614067
(1)
|
1 / 2 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | CHEMBL11298 |
CHEMBL1741322
(3)
|
0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | CHEMBL11298 |
CHEMBL1741323
(3)
|
1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL11298 |
CHEMBL1741324
(3)
|
0 / 1 |
| Q7Z2H8 | Proton-coupled amino acid transporter 1 | Unclassified protein | CHEMBL11298 CHEMBL285123 |
CHEMBL1919336
(4)
|
0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | CHEMBL11298 |
CHEMBL1794536
(1)
|
0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | CHEMBL11298 |
CHEMBL1613914
(2)
|
0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | CHEMBL285123 |
CHEMBL1738442
(2)
|
0 / 0 |
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (18)
| OMIM | preferred title | UniProt |
|---|---|---|
| #210900 | Bloom syndrome; blm |
P54132
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
KEGG DISEASE (17)
| KEGG | disease name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|