Metabolite
KNApSAcK Entry
| id | C00007562 |
|---|---|
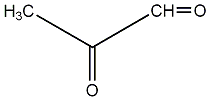
| Name | Methylglyoxal |
| CAS RN | 78-98-8 |
| Standard InChI | InChI=1S/C3H4O2/c1-3(5)2-4/h2H,1H3 |
| Standard InChI (Main Layer) | InChI=1S/C3H4O2/c1-3(5)2-4/h2H,1H3 |
Cluster
| Phytochemical cluster | |
|---|---|
| KCF-S cluster | No. 7330 |
Link
ChEMBL
| By standard InChI | CHEMBL170721 |
|---|---|
| By standard InChI Main Layer | CHEMBL170721 |
KEGG
| By LinkDB | C00546 |
|---|
CTD
| By CAS RN | D011765 |
|---|
Species
Summary
Plant class
| class name | count |
|---|
Family
| family name | count |
|---|---|
| Enterobacteriaceae | 1 |
List (1)
* NCBI
| KNApSAcK organism | *ID | *family | *plant class | *kingdom |
|---|---|---|---|---|
| Escherichia coli | 562 | Enterobacteriaceae | Bacteria |
Human Protein / Gene in interaction
CTD interaction (26)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D011765 | 10327 |
AKR1A1
ALDR1 ALR ARM DD3 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) (EC:1.1.1.2) | AKR1A1 protein results in increased reduction of Pyruvaldehyde |
increases reduction
|
protein |
10510318
|
| D011765 | 231 |
AKR1B1
ADR ALDR1 ALR2 AR |
aldo-keto reductase family 1, member B1 (aldose reductase) (EC:1.1.1.21) | AKR1B1 protein results in increased reduction of Pyruvaldehyde |
increases reduction
|
protein |
10510318
|
| D011765 | 8574 |
AKR7A2
AFAR AFAR1 AFB1-AR1 AKR7 |
aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) (EC:1.1.1.n11) | AKR7A2 protein results in decreased susceptibility to Pyruvaldehyde |
decreases response to substance
|
protein |
22001351
|
| D011765 | 213 |
ALB
PRO0883 PRO0903 PRO1341 |
albumin | Pyruvaldehyde binds to ALB protein |
affects binding
|
protein |
20934417
|
| D011765 | 213 |
ALB
PRO0883 PRO0903 PRO1341 |
albumin | [Pyruvaldehyde binds to ALB protein] which results in decreased metabolism of Prostaglandins |
affects binding
/ decreases metabolic processing |
protein |
20934417
|
| D011765 | 213 |
ALB
PRO0883 PRO0903 PRO1341 |
albumin | Pyruvaldehyde inhibits the reaction [Warfarin binds to ALB protein] |
affects binding
/ decreases reaction |
protein |
20934417
|
| D011765 | 836 |
CASP3
CPP32 CPP32B SCA-1 |
caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) | Pyruvaldehyde results in increased cleavage of CASP3 protein |
increases cleavage
|
protein |
18565324
|
| D011765 | 836 |
CASP3
CPP32 CPP32B SCA-1 |
caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) | telmisartan inhibits the reaction [Pyruvaldehyde results in increased cleavage of CASP3 protein] |
decreases reaction
/ increases cleavage |
protein |
18565324
|
| D011765 | 1356 |
CP
CP-2 |
ceruloplasmin (ferroxidase) (EC:1.16.3.1) | bathocuproine inhibits the reaction [Pyruvaldehyde promotes the reaction [CP protein binds to CP protein]] |
affects binding
/ decreases reaction / increases reaction |
protein |
16756764
|
| D011765 | 1356 |
CP
CP-2 |
ceruloplasmin (ferroxidase) (EC:1.16.3.1) | Ditiocarb inhibits the reaction [Pyruvaldehyde promotes the reaction [CP protein binds to CP protein]] |
affects binding
/ decreases reaction / increases reaction |
protein |
16756764
|
| D011765 | 1356 |
CP
CP-2 |
ceruloplasmin (ferroxidase) (EC:1.16.3.1) | formic acid inhibits the reaction [Pyruvaldehyde promotes the reaction [CP protein binds to CP protein]] |
affects binding
/ decreases reaction / increases reaction |
protein |
16756764
|
| D011765 | 1356 |
CP
CP-2 |
ceruloplasmin (ferroxidase) (EC:1.16.3.1) | Pyruvaldehyde promotes the reaction [CP protein binds to CP protein] |
affects binding
/ increases reaction |
protein |
16756764
|
| D011765 | 1356 |
CP
CP-2 |
ceruloplasmin (ferroxidase) (EC:1.16.3.1) | Pyruvaldehyde results in decreased activity of CP protein |
decreases activity
|
protein |
16756764
|
| D011765 | 1356 |
CP
CP-2 |
ceruloplasmin (ferroxidase) (EC:1.16.3.1) | Sodium Azide inhibits the reaction [Pyruvaldehyde promotes the reaction [CP protein binds to CP protein]] |
affects binding
/ decreases reaction / increases reaction |
protein |
16756764
|
| D011765 | 1520 |
CTSS
|
cathepsin S (EC:3.4.22.27) | Pyruvaldehyde results in decreased activity of CTSS protein |
decreases activity
|
protein |
16671891
|
| D011765 | 2739 |
GLO1
GLOD1 GLYI |
glyoxalase I (EC:4.4.1.5) | GLO1 protein affects the reaction [Pyruvaldehyde binds to HSPB1 protein] |
affects binding
/ affects reaction |
protein |
20093988
|
| D011765 | 2739 |
GLO1
GLOD1 GLYI |
glyoxalase I (EC:4.4.1.5) | GLO1 protein affects the susceptibility to Pyruvaldehyde |
affects response to substance
|
protein |
20093988
|
| D011765 | 3315 |
HSPB1
CMT2F HMN2B HS.76067 HSP27 HSP28 Hsp25 SRP27 |
heat shock 27kDa protein 1 | GLO1 protein affects the reaction [Pyruvaldehyde binds to HSPB1 protein] |
affects binding
/ affects reaction |
protein |
20093988
|
| D011765 | 3315 |
HSPB1
CMT2F HMN2B HS.76067 HSP27 HSP28 Hsp25 SRP27 |
heat shock 27kDa protein 1 | Pyruvaldehyde binds to HSPB1 protein |
affects binding
|
protein |
20093988
|
| D011765 | 3569 |
IL6
BSF2 HGF HSF IFNB2 IL-6 |
interleukin 6 (interferon, beta 2) | Pyruvaldehyde analog affects the expression of IL6 mRNA |
affects expression
|
mRNA |
20200221
|
| D011765 | 3576 |
IL8
CXCL8 GCP-1 GCP1 LECT LUCT LYNAP MDNCF MONAP NAF NAP-1 NAP1 |
interleukin 8 | Pyruvaldehyde analog affects the expression of IL8 mRNA |
affects expression
|
mRNA |
20200221
|
| D011765 | 5444 |
PON1
ESA MVCD5 PON |
paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) | Pyruvaldehyde inhibits the reaction [PON1 protein results in increased hydrolysis of Paraoxon] |
decreases reaction
/ increases hydrolysis |
protein |
15375178
|
| D011765 | 5444 |
PON1
ESA MVCD5 PON |
paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) | Pyruvaldehyde inhibits the reaction [PON1 protein results in increased hydrolysis of phenylacetic acid] |
decreases reaction
/ increases hydrolysis |
protein |
15375178
|
| D011765 | 5444 |
PON1
ESA MVCD5 PON |
paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) | Pyruvaldehyde results in decreased activity of PON1 protein |
decreases activity
|
protein |
15375178
|
| D011765 | 8989 |
TRPA1
ANKTM1 FEPS |
transient receptor potential cation channel, subfamily A, member 1 | Pyruvaldehyde results in increased activity of TRPA1 protein |
increases activity
|
protein |
22863862
|
| D011765 | 8989 |
TRPA1
ANKTM1 FEPS |
transient receptor potential cation channel, subfamily A, member 1 | [Pyruvaldehyde results in increased activity of TRPA1 protein] which results in increased transport of Calcium |
increases activity
/ increases transport |
protein |
22863862
|
Related Disease
Diseases related to CTD interactions
11 disease from interactions of metabolites
| MeSH disease | OMIM | compound | disease name | evidence type |
reference
pmid |
|---|---|---|---|---|---|
| D003920 | D011765 | Diabetes Mellitus |
marker/mechanism
|
11559049
11719831 18729331 |
|
| D058065 | D011765 | Diabetic Cardiomyopathies |
marker/mechanism
|
20621070
|
|
| D003928 | D011765 | Diabetic Nephropathies |
marker/mechanism
|
20621070
|
|
| D003930 | D011765 | Diabetic Retinopathy |
marker/mechanism
|
20621070
|
|
| D056486 | D011765 | Drug-Induced Liver Injury |
marker/mechanism
|
20211157
|
|
| D006943 | D011765 | Hyperglycemia |
marker/mechanism
|
11719831
|
|
| D006967 | D011765 | Hypersensitivity |
marker/mechanism
|
17347135
|
|
| D007249 | D011765 | Inflammation |
marker/mechanism
|
17347135
22101032 |
|
| D009336 | D011765 | Necrosis |
marker/mechanism
|
11331073
|
|
| D056627 | D011765 | Peritoneal Fibrosis |
marker/mechanism
|
22101032
|
|
| D011230 | D011765 | Precancerous Conditions |
marker/mechanism
|
7821871
|