Elaeagnus multiflora
Species
KNApSAcK Entry
| Organism name | Elaeagnus multiflora |
|---|---|
| Genus | Elaeagnus |
| Family | Elaeagnaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Elaeagnus multiflora |
|---|---|
| Linked NCBI taxonomy ID | 99570 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Elaeagnaceae |
|---|---|
| ID | 25996 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (5)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
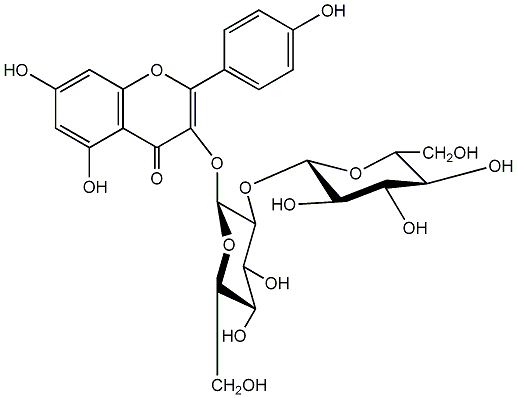
C00005165
|
Sophoraflavonoloside
/ Kaempferol 3-O-sophoroside / Kaempferol 3-O-beta-sophoroside |
CHEMBL500233
CHEMBL1375795 CHEMBL2032411 |
C064309
|
12 / 2 / 2 | 6 / 4 | No. 1 | No. 15 |
|
|
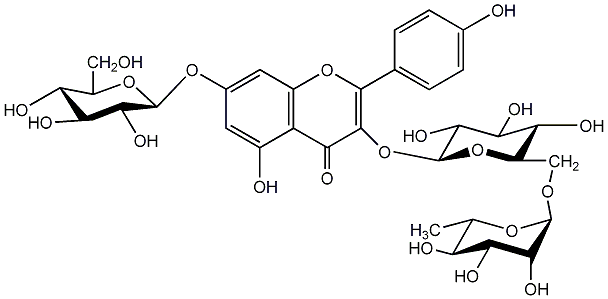
C00005239
|
Kaempferol 3-rutinoside-7-glucoside
|
No. 5 | No. 15 |
|
||||
|
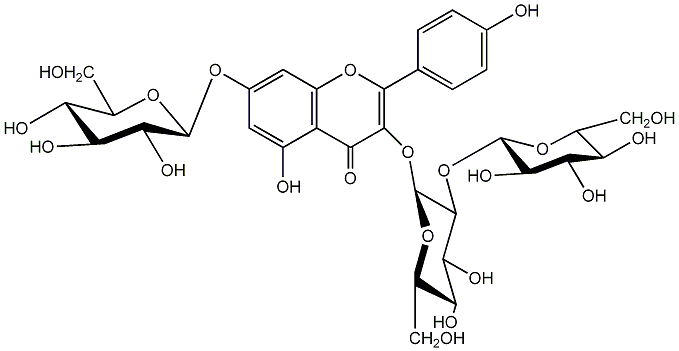
C00005229
|
(-)-Kaempferol 3-O-sophoroside 7-O-beta-D-glucopyranoside
/ Kaempferol 3-O-sophoroside 7-O-beta-D-glucopyranoside:Kaempferol 3-sophoroside-7-glucoside |
No. 5 | No. 15 |
|
||||
|
C00005248
|
Kaempferol 3-glucoside-7-sophoroside
|
No. 5 | No. 15 |

|
||||
|
C00005579
|
Isorhamnetin 3-sophoroside-7-glucoside
|
No. 5 | No. 15 |

|
Human Protein / Gene in interactions
12 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P06746 | DNA polymerase beta | Enzyme | C00005165 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00005165 | 1 / 1 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00005165 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00005165 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00005165 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00005165 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00005165 | 1 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00005165 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00005165 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00005165 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00005165 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00005165 | 0 / 0 |
6 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00005165
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00005165
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00005165
|
| 7097 | TLR2, CD282, TIL4 | toll-like receptor 2 |
C00005165
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00005165
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00005165
|