Mildbraediodendron excelsum
Species
KNApSAcK Entry
| Organism name | Mildbraediodendron excelsum |
|---|---|
| Genus | Mildbraediodendron |
| Family | Fabaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Mildbraediodendron excelsum |
|---|---|
| Linked NCBI taxonomy ID | 149663 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Fabaceae |
|---|---|
| ID | 3803 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (7)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
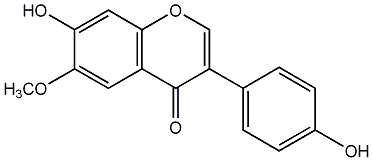
C00009392
|
Glycitein
/ 7,4'-Dihydroxy-6-methoxyisoflavone |
CHEMBL513024
|
C086566
|
2 / 2 / 2 | 40 / 4 | No. 3 | No. 15 |
|
|
C00009428
|
6,7,2',4',5'-Pentamethoxyisoflavone
|
CHEMBL234933
|
No. 8 | No. 15 |

|
|||
|
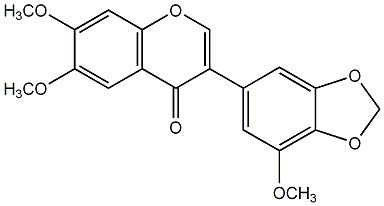
C00009422
|
6,7,3'-Trimethoxy-4',5'-methylenedioxyisoflavone
|
No. 27 | No. 15 |
|
||||
|
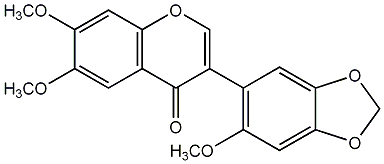
C00009421
|
Milldurone
/ 6,7,2'-Trimethoxy-4',5'-methylenedioxyisoflavone |
CHEMBL234717
|
No. 27 | No. 15 |
|
|||
|
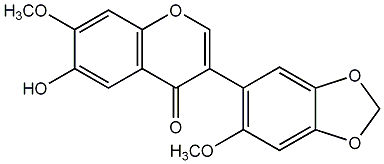
C00009418
|
6-Hydroxy-7,2'-dimethoxy-4',5'-methylenedioxyisoflavone
|
No. 27 | No. 15 |
|
||||
|
C00020211
|
6,7-Dihydroxy-3',4'-methylenedioxyisoflavone
|
CHEMBL239030
|
3 / 2 / 0 | No. 27 | No. 15 |

|
||
|
C00009544
|
6,7-Dimethoxy-3',4'-methylenedioxyisoflavanone
|
No. 77 | No. 14 |

|
Human Protein / Gene in interactions
5 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00009392 | 1 / 1 |
| O15296 | Arachidonate 15-lipoxygenase B | Enzyme | C00020211 | 0 / 0 |
| P18054 | Arachidonate 12-lipoxygenase, 12S-type | Enzyme | C00020211 | 2 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00009392 | 1 / 1 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00020211 | 0 / 0 |
40 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 19 | ABCA1, ABC-1, ABC1, CERP, HDLDT1, TGD | ATP-binding cassette, sub-family A (ABC1), member 1 |
C00009392
|
| 119 | ADD2, ADDB | adducin 2 (beta) |
C00009392
|
| 396 | ARHGDIA, GDIA1, NPHS8, RHOGDI, RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha |
C00009392
|
| 23545 | ATP6V0A2, A2, ARCL, ARCL2A, ATP6A2, ATP6N1D, J6B7, RTF, STV1, TJ6, TJ6M, TJ6S, VPH1, WSS | ATPase, H+ transporting, lysosomal V0 subunit a2 (EC:3.6.3.6) |
C00009392
|
| 11177 | BAZ1A, ACF1, WALp1, WCRF180, hACF1 | bromodomain adjacent to zinc finger domain, 1A |
C00009392
|
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00009392
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00009392
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00009392
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00009392
|
| 1795 | DOCK3, MOCA, PBP | dedicator of cytokinesis 3 |
C00009392
|
| 2041 | EPHA1, EPH, EPHT, EPHT1 | EPH receptor A1 (EC:2.7.10.1) |
C00009392
|
| 83715 | ESPN, DFNB36 | espin |
C00009392
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00009392
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00009392
|
| 56776 | FMN2 | formin 2 |
C00009392
|
| 7866 | IFRD2, IFNRP, SKMc15, SM15 | interferon-related developmental regulator 2 |
C00009392
|
| 26145 | IRF2BP1 | interferon regulatory factor 2 binding protein 1 |
C00009392
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00009392
|
| 26251 | KCNG2, KCNF2, KV6.2 | potassium voltage-gated channel, subfamily G, member 2 |
C00009392
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00009392
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00009392
|
| 4314 | MMP3, CHDS6, MMP-3, SL-1, STMY, STMY1, STR1 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) (EC:3.4.24.17) |
C00009392
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00009392
|
| 8204 | NRIP1, RIP140 | nuclear receptor interacting protein 1 |
C00009392
|
| 56944 | OLFML3, HNOEL-iso, OLF44 | olfactomedin-like 3 |
C00009392
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00009392
|
| 64236 | PDLIM2, MYSTIQUE, SLIM | PDZ and LIM domain 2 (mystique) |
C00009392
|
| 5411 | PNN, DRS, DRSP, SDK3, memA | pinin, desmosome associated protein |
C00009392
|
| 5458 | POU4F2, BRN3.2, BRN3B, Brn-3b | POU class 4 homeobox 2 |
C00009392
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00009392
|
| 81537 | SGPP1, SPPase1 | sphingosine-1-phosphate phosphatase 1 |
C00009392
|
| 6558 | SLC12A2, BSC, BSC2, NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
C00009392
|
| 6667 | SP1 | Sp1 transcription factor |
C00009392
|
| 6781 | STC1, STC | stanniocalcin 1 |
C00009392
|
| 6870 | TACR3, HH11, NK-3R, NK3R, NKR, TAC3RL | tachykinin receptor 3 |
C00009392
|
| 7130 | TNFAIP6, TSG-6, TSG6 | tumor necrosis factor, alpha-induced protein 6 |
C00009392
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00009392
|
| 9537 | TP53I11, PIG11 | tumor protein p53 inducible protein 11 |
C00009392
|
| 7345 | UCHL1, NDGOA, PARK5, PGP_9.5, PGP9.5, PGP95, Uch-L1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (EC:3.4.19.12) |
C00009392
|
| 7421 | VDR, NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
C00009392
|