Streptomyces venezuelae
Species
KNApSAcK Entry
| Organism name | Streptomyces venezuelae |
|---|---|
| Genus | Streptomyces |
| Family | Streptomycetaceae |
| Kingdom | Bacteria |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Streptomyces venezuelae |
|---|---|
| Linked NCBI taxonomy ID | 54571 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Streptomycetaceae |
|---|---|
| ID | 2062 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Bacteria |
|---|---|
| ID | 2 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (5)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00044988
|
p-(Acetylamino)benzyl alcohol
|
C057933
|
No. 4605 |
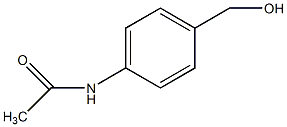
|
||||
|
C00044491
|
Acetaminophen
|
CHEMBL112
|
D000082
|
132 / 57 / 47 | 4130 / 166 | No. 4605 |
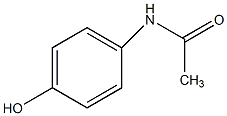
|
|
|
C00044987
|
p-(Acetylamino)benzoic acid
|
CHEMBL112687
|
C023730
|
6 / 6 / 6 | No. 8182 |
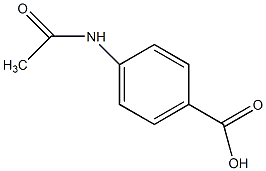
|
||
|
C00016835
|
Jadomycin B
|
CHEMBL1631684
|
C081750
|
No. 8616 |
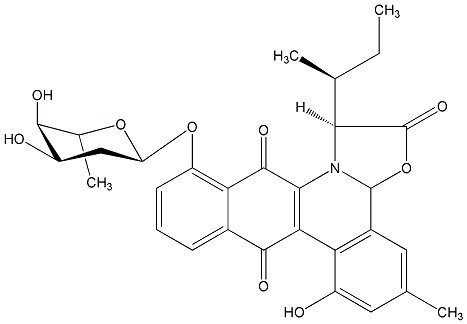
|
|||
|
C00044658
|
Corynecin II
|
CHEMBL25253
|
No. 8658 |

|
Human Protein / Gene in interactions
135 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00044491 C00044987 | 3 / 2 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00044491 C00044987 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00044491 C00044987 | 0 / 0 |
| P36537 | UDP-glucuronosyltransferase 2B10 | Enzyme | C00044491 | 0 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00044491 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00044491 | 0 / 3 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00044491 | 2 / 2 |
| P08246 | Neutrophil elastase | S1A | C00044491 | 2 / 1 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00044491 | 0 / 0 |
| Q16539 | Mitogen-activated protein kinase 14 | p38 | C00044491 | 0 / 0 |
| P49146 | Neuropeptide Y receptor type 2 | Neuropeptide Y receptor | C00044491 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00044491 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00044491 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00044491 | 0 / 0 |
| P29466 | Caspase-1 | C14 | C00044491 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00044491 | 0 / 0 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00044491 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00044491 | 2 / 2 |
| O15245 | Solute carrier family 22 member 1 | Drug uniporter | C00044491 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00044491 | 1 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00044491 | 1 / 2 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00044491 | 0 / 1 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00044491 | 1 / 1 |
| P25021 | Histamine H2 receptor | Histamine receptor | C00044491 | 0 / 0 |
| P35367 | Histamine H1 receptor | Histamine receptor | C00044491 | 0 / 0 |
| Q01959 | Sodium-dependent dopamine transporter | Dopamine | C00044491 | 1 / 0 |
| P08912 | Muscarinic acetylcholine receptor M5 | Acetylcholine receptor | C00044491 | 0 / 0 |
| P18825 | Alpha-2C adrenergic receptor | Adrenergic receptor | C00044491 | 0 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00044491 | 0 / 0 |
| P02768 | Serum albumin | Secreted protein | C00044491 | 0 / 0 |
| P04798 | Cytochrome P450 1A1 | Cytochrome P450 1A1 | C00044491 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00044491 | 1 / 0 |
| P25024 | C-X-C chemokine receptor type 1 | CXC chemokine receptor | C00044491 | 0 / 0 |
| Q9UQ49 | Sialidase-3 | Enzyme | C00044987 | 0 / 0 |
| P06241 | Tyrosine-protein kinase Fyn | Src | C00044491 | 0 / 0 |
| Q08209 | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ser_Thr | C00044491 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00044491 | 0 / 1 |
| P16662 | UDP-glucuronosyltransferase 2B7 | Enzyme | C00044491 | 0 / 0 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00044491 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00044491 | 1 / 8 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | C00044491 | 2 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00044491 | 0 / 0 |
| P37288 | Vasopressin V1a receptor | Vasopressin and oxytocin receptor | C00044491 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00044491 | 0 / 0 |
| Q9Y271 | Cysteinyl leukotriene receptor 1 | Leukotriene receptor | C00044491 | 0 / 0 |
| P29274 | Adenosine receptor A2a | Adenosine receptor | C00044491 | 0 / 0 |
| P34972 | Cannabinoid receptor 2 | Cannabinoid receptor | C00044491 | 0 / 0 |
| P20292 | Arachidonate 5-lipoxygenase-activating protein | Other cytosolic protein | C00044491 | 1 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00044491 | 0 / 0 |
| P25929 | Neuropeptide Y receptor type 1 | Neuropeptide Y receptor | C00044491 | 0 / 0 |
| P50052 | Type-2 angiotensin II receptor | Angiotensin receptor | C00044491 | 1 / 1 |
| P17948 | Vascular endothelial growth factor receptor 1 | Vegfr | C00044491 | 0 / 0 |
| P41968 | Melanocortin receptor 3 | Melanocortin receptor | C00044491 | 1 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00044491 | 0 / 0 |
| P25440 | Bromodomain-containing protein 2 | Unclassified protein | C00044491 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00044491 | 1 / 2 |
| P11509 | Cytochrome P450 2A6 | Cytochrome P450 2A6 | C00044491 | 0 / 0 |
| P42330 | Aldo-keto reductase family 1 member C3 | Enzyme | C00044491 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00044491 | 2 / 0 |
| P50225 | Sulfotransferase 1A1 | Enzyme | C00044491 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00044491 | 0 / 0 |
| P54855 | UDP-glucuronosyltransferase 2B15 | Enzyme | C00044491 | 0 / 0 |
| P04035 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Oxidoreductase | C00044491 | 0 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00044491 | 0 / 0 |
| P21917 | D(4) dopamine receptor | Dopamine receptor | C00044491 | 0 / 0 |
| P30988 | Calcitonin receptor | Calcitonin receptor | C00044491 | 0 / 0 |
| P35462 | D(3) dopamine receptor | Dopamine receptor | C00044491 | 1 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00044491 | 0 / 0 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00044491 | 0 / 1 |
| P41595 | 5-hydroxytryptamine receptor 2B | Serotonin receptor | C00044491 | 0 / 0 |
| P25101 | Endothelin-1 receptor | Endothelin receptor | C00044491 | 0 / 0 |
| P30411 | B2 bradykinin receptor | Bradykinin receptor | C00044491 | 0 / 0 |
| P32245 | Melanocortin receptor 4 | Melanocortin receptor | C00044491 | 1 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00044491 | 0 / 0 |
| P32238 | Cholecystokinin receptor type A | Cholecystokinin receptor | C00044491 | 0 / 0 |
| P08311 | Cathepsin G | S1A | C00044491 | 0 / 0 |
| Q99720 | Sigma non-opioid intracellular receptor 1 | Membrane receptor | C00044491 | 1 / 0 |
| P03956 | Interstitial collagenase | M10A | C00044491 | 0 / 1 |
| P32241 | Vasoactive intestinal polypeptide receptor 1 | Vasoactive intestinal peptide receptor | C00044491 | 0 / 0 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00044491 | 3 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00044491 | 0 / 1 |
| P08172 | Muscarinic acetylcholine receptor M2 | Acetylcholine receptor | C00044491 | 2 / 0 |
| P11229 | Muscarinic acetylcholine receptor M1 | Acetylcholine receptor | C00044491 | 0 / 0 |
| P21554 | Cannabinoid receptor 1 | Cannabinoid receptor | C00044491 | 0 / 0 |
| P31645 | Sodium-dependent serotonin transporter | Serotonin | C00044491 | 2 / 0 |
| P04626 | Receptor tyrosine-protein kinase erbB-2 | TK tyrosine-protein kinase EGFR subfamily | C00044491 | 5 / 9 |
| P20309 | Muscarinic acetylcholine receptor M3 | Acetylcholine receptor | C00044491 | 1 / 0 |
| P21452 | Substance-K receptor | Neurokinin receptor | C00044491 | 0 / 0 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00044491 | 0 / 0 |
| P51681 | C-C chemokine receptor type 5 | CC chemokine receptor | C00044491 | 3 / 0 |
| P50406 | 5-hydroxytryptamine receptor 6 | Serotonin receptor | C00044491 | 0 / 0 |
| P41597 | C-C chemokine receptor type 2 | CC chemokine receptor | C00044491 | 1 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00044491 | 0 / 0 |
| P08575 | Receptor-type tyrosine-protein phosphatase C | Enzyme | C00044491 | 2 / 1 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00044491 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00044491 | 0 / 1 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | C00044491 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00044491 | 0 / 0 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00044491 | 0 / 0 |
| P06133 | UDP-glucuronosyltransferase 2B4 | Enzyme | C00044491 | 0 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00044491 | 0 / 0 |
| O76074 | cGMP-specific 3',5'-cyclic phosphodiesterase | PDE_5A | C00044491 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00044491 | 1 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00044491 | 1 / 0 |
| P22303 | Acetylcholinesterase | Hydrolase | C00044491 | 1 / 0 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | C00044491 | 0 / 0 |
| P28335 | 5-hydroxytryptamine receptor 2C | Serotonin receptor | C00044491 | 0 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00044491 | 0 / 0 |
| P08173 | Muscarinic acetylcholine receptor M4 | Acetylcholine receptor | C00044491 | 0 / 0 |
| P25103 | Substance-P receptor | Neurokinin receptor | C00044491 | 0 / 0 |
| P25105 | Platelet-activating factor receptor | PAF receptor | C00044491 | 0 / 0 |
| P33032 | Melanocortin receptor 5 | Melanocortin receptor | C00044491 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00044491 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00044491 | 0 / 1 |
| P05181 | Cytochrome P450 2E1 | Cytochrome P450 2E1 | C00044491 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00044987 | 0 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00044491 | 5 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00044491 | 4 / 3 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00044491 | 0 / 0 |
| Q15059 | Bromodomain-containing protein 3 | Unclassified protein | C00044491 | 0 / 0 |
| O60885 | Bromodomain-containing protein 4 | Unclassified protein | C00044491 | 0 / 0 |
| P10275 | Androgen receptor | NR3C4 | C00044987 | 3 / 4 |
| P23975 | Sodium-dependent noradrenaline transporter | Norepinephrine | C00044491 | 1 / 1 |
| P25100 | Alpha-1D adrenergic receptor | Adrenergic receptor | C00044491 | 0 / 0 |
| P30542 | Adenosine receptor A1 | Adenosine receptor | C00044491 | 0 / 0 |
| P18089 | Alpha-2B adrenergic receptor | Adrenergic receptor | C00044491 | 0 / 0 |
| P24557 | Thromboxane-A synthase | Cytochrome P450 5A1 | C00044491 | 1 / 1 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00044491 | 0 / 0 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00044491 | 0 / 1 |
| P25025 | C-X-C chemokine receptor type 2 | CXC chemokine receptor | C00044491 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00044491 | 1 / 1 |
| Q99685 | Monoglyceride lipase | Enzyme | C00044491 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00044491 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00044491 | 1 / 2 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00044491 | 0 / 0 |
4130 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 127550 | A3GALT2, A3GALT2P, IGBS3S | alpha 1,3-galactosyltransferase 2 |
C00044491
|
| 22848 | AAK1 | AP2 associated kinase 1 (EC:2.7.11.1) |
C00044491
|
| 441376 | AARD, C8orf85 | alanine and arginine rich domain containing protein |
C00044491
|
| 60496 | AASDHPPT, AASD-PPT, LYS2, LYS5 | aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (EC:2.7.8.-) |
C00044491
|
| 10157 | AASS, LKR/SDH, LKRSDH, LORSDH | aminoadipate-semialdehyde synthase (EC:1.5.1.8 1.5.1.9) |
C00044491
|
| 19 | ABCA1, ABC-1, ABC1, CERP, HDLDT1, TGD | ATP-binding cassette, sub-family A (ABC1), member 1 |
C00044491
|
| 23460 | ABCA6, EST155051 | ATP-binding cassette, sub-family A (ABC1), member 6 |
C00044491
|
| 10351 | ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 |
C00044491
|
| 10350 | ABCA9, EST640918 | ATP-binding cassette, sub-family A (ABC1), member 9 |
C00044491
|
| 5243 | ABCB1, ABC20, CD243, CLCS, GP170, MDR1, P-GP, PGY1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 (EC:3.6.3.44) |
C00044491
|
| 8647 | ABCB11, ABC16, BRIC2, BSEP, PFIC-2, PFIC2, PGY4, SPGP | ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
C00044491
|
| 5244 | ABCB4, ABC21, GBD1, ICP3, MDR2, MDR2/3, MDR3, PFIC-3, PGY3 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 (EC:3.6.3.44) |
C00044491
|
| 10058 | ABCB6, ABC, ABC14, DUH3, LAN, MCOPCB7, MTABC3, PRP, umat | ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
C00044491
|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00044491
|
| 1244 | ABCC2, ABC30, CMOAT, DJS, MRP2, cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
C00044491
|
| 8714 | ABCC3, ABC31, EST90757, MLP2, MOAT-D, MRP3, cMOAT2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
C00044491
|
| 10257 | ABCC4, EST170205, MOAT-B, MOATB, MRP4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
C00044491
|
| 10057 | ABCC5, ABC33, EST277145, MOAT-C, MOATC, MRP5, SMRP, pABC11 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
C00044491
|
| 368 | ABCC6, ABC34, ARA, EST349056, GACI2, MLP1, MOAT-E, MOATE, MRP6, PXE, PXE1, URG7 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
C00044491
|
| 215 | ABCD1, ABC42, ALD, ALDP, AMN | ATP-binding cassette, sub-family D (ALD), member 1 |
C00044491
|
| 6059 | ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I | ATP-binding cassette, sub-family E (OABP), member 1 |
C00044491
|
| 9429 | ABCG2, ABC15, ABCP, BCRP, BCRP1, BMDP, CD338, CDw338, EST157481, GOUT1, MRX, MXR, MXR1, UAQTL1 | ATP-binding cassette, sub-family G (WHITE), member 2 |
C00044491
|
| 64240 | ABCG5, STSL | ATP-binding cassette, sub-family G (WHITE), member 5 |
C00044491
|
| 26090 | ABHD12, ABHD12A, BEM46L2, C20orf22, PHARC, dJ965G21.2 | abhydrolase domain containing 12 (EC:3.1.1.23) |
C00044491
|
| 7920 | ABHD16A, BAT5, D6S82E, NG26, PP199 | abhydrolase domain containing 16A |
C00044491
|
| 140701 | ABHD16B, C20orf135, dJ591C20.1 | abhydrolase domain containing 16B |
C00044491
|
| 51104 | ABHD17B, C9orf77, FAM108B1, RP11-409O11.2 | abhydrolase domain containing 17B |
C00044491
|
| 58489 | ABHD17C, FAM108C1 | abhydrolase domain containing 17C |
C00044491
|
| 11057 | ABHD2, HS1-2, LABH2, PHPS1-2 | abhydrolase domain containing 2 |
C00044491
|
| 51099 | ABHD5, CDS, CGI58, IECN2, NCIE2 | abhydrolase domain containing 5 (EC:2.3.1.51) |
C00044491
|
| 10152 | ABI2, ABI-2, ABI2B, AIP-1, AblBP3, SSH3BP2, argBP1, argBPIA, argBPIB | abl-interactor 2 |
C00044491
|
| 3983 | ABLIM1, ABLIM, LIMAB1, LIMATIN, abLIM-1 | actin binding LIM protein 1 |
C00044491
|
| 84448 | ABLIM2 | actin binding LIM protein family, member 2 |
C00044491
|
| 22885 | ABLIM3 | actin binding LIM protein family, member 3 |
C00044491
|
| 28 | ABO, A3GALNT, A3GALT1, GTB, NAGAT | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (EC:2.4.1.37 2.4.1.40) |
C00044491
|
| 25841 | ABTB2, ABTB2A, BTBD22 | ankyrin repeat and BTB (POZ) domain containing 2 |
C00044491
|
| 30 | ACAA1, ACAA, PTHIO, THIO | acetyl-CoA acyltransferase 1 (EC:2.3.1.16) |
C00044491
|
| 32 | ACACB, ACC2, ACCB, HACC275 | acetyl-CoA carboxylase beta (EC:6.4.1.2 6.3.4.14) |
C00044491
|
| 23527 | ACAP2, CENTB2, CNT-B2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
C00044491
|
| 116983 | ACAP3, CENTB5 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
C00044491
|
| 39 | ACAT2 | acetyl-CoA acetyltransferase 2 (EC:2.3.1.9) |
C00044491
|
| 91452 | ACBD5 | acyl-CoA binding domain containing 5 |
C00044491
|
| 414149 | ACBD7, bA455B2.2 | acyl-CoA binding domain containing 7 |
C00044491
|
| 65057 | ACD, PIP1, PTOP, TINT1, TPP1 | adrenocortical dysplasia homolog (mouse) |
C00044491
|
| 340485 | ACER2, ALKCDase2, ASAH3L | alkaline ceramidase 2 (EC:3.5.1.23) |
C00044491
|
| 55331 | ACER3, APHC, PHCA | alkaline ceramidase 3 (EC:3.5.1.23) |
C00044491
|
| 22985 | ACIN1, ACINUS, ACN, fSAP152 | apoptotic chromatin condensation inducer 1 |
C00044491
|
| 57007 | ACKR3, CMKOR1, CXC-R7, CXCR-7, CXCR7, GPR159, RDC-1, RDC1 | atypical chemokine receptor 3 |
C00044491
|
| 641371 | ACOT1, ACH2, CTE-1, LACH2 | acyl-CoA thioesterase 1 (EC:3.1.2.2) |
C00044491
|
| 10965 | ACOT2, CTE-IA, CTE1A, MTE1, PTE2, PTE2A, ZAP128 | acyl-CoA thioesterase 2 (EC:3.1.2.2) |
C00044491
|
| 51 | ACOX1, ACOX, PALMCOX, SCOX | acyl-CoA oxidase 1, palmitoyl (EC:1.3.3.6) |
C00044491
|
| 8310 | ACOX3 | acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6) |
C00044491
|
| 92370 | ACPL2 | acid phosphatase-like 2 (EC:3.1.3.2) |
C00044491
|
| 49 | ACR | acrosin (EC:3.4.21.10) |
C00044491
|
| 23205 | ACSBG1, BG, BG1, BGM, GR-LACS, LPD | acyl-CoA synthetase bubblegum family member 1 (EC:6.2.1.3) |
C00044491
|
| 2180 | ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2 | acyl-CoA synthetase long-chain family member 1 (EC:6.2.1.3) |
C00044491
|
| 2181 | ACSL3, ACS3, FACL3, PRO2194 | acyl-CoA synthetase long-chain family member 3 (EC:6.2.1.3) |
C00044491
|
| 51703 | ACSL5, ACS2, ACS5, FACL5 | acyl-CoA synthetase long-chain family member 5 (EC:6.2.1.3) |
C00044491
|
| 123876 | ACSM2A, A-923A4.1, ACSM2 | acyl-CoA synthetase medium-chain family member 2A (EC:6.2.1.2) |
C00044491
|
| 79611 | ACSS3 | acyl-CoA synthetase short-chain family member 3 (EC:6.2.1.1) |
C00044491
|
| 59 | ACTA2, AAT6, ACTSA, MYMY5 | actin, alpha 2, smooth muscle, aorta |
C00044491
|
| 71 | ACTG1, ACT, ACTG, BRWS2, DFNA20, DFNA26 | actin, gamma 1 |
C00044491
|
| 87 | ACTN1, BDPLT15 | actinin, alpha 1 |
C00044491
|
| 88 | ACTN2, CMD1AA | actinin, alpha 2 |
C00044491
|
| 10096 | ACTR3, ARP3 | ARP3 actin-related protein 3 homolog (yeast) |
C00044491
|
| 84517 | ACTRT3, ARP-T3, ARPM1 | actin-related protein T3 |
C00044491
|
| 91 | ACVR1B, ACTRIB, ACVRLK4, ALK4, SKR2 | activin A receptor, type IB (EC:2.7.11.30) |
C00044491
|
| 92 | ACVR2A, ACTRII, ACVR2 | activin A receptor, type IIA (EC:2.7.11.30) |
C00044491
|
| 102 | ADAM10, AD10, CD156c, HsT18717, MADM, kuz | ADAM metallopeptidase domain 10 (EC:3.4.24.81) |
C00044491
|
| 8751 | ADAM15, MDC15 | ADAM metallopeptidase domain 15 |
C00044491
|
| 8728 | ADAM19, MADDAM, MLTNB | ADAM metallopeptidase domain 19 |
C00044491
|
| 80332 | ADAM33, C20orf153, DJ964F7.1 | ADAM metallopeptidase domain 33 |
C00044491
|
| 81794 | ADAMTS10, ADAM-TS10, ADAMTS-10, WMS, WMS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
C00044491
|
| 81792 | ADAMTS12, PRO4389 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
C00044491
|
| 9507 | ADAMTS4, ADAMTS-2, ADAMTS-4, ADMP-1 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 (EC:3.4.24.82) |
C00044491
|
| 11096 | ADAMTS5, ADAM-TS_11, ADAM-TS_5, ADAM-TS5, ADAMTS-11, ADAMTS-5, ADAMTS11, ADMP-2 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
C00044491
|
| 11095 | ADAMTS8, ADAM-TS8, METH2 | ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
C00044491
|
| 56999 | ADAMTS9 | ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
C00044491
|
| 57188 | ADAMTSL3 | ADAMTS-like 3 |
C00044491
|
| 103 | ADAR, ADAR1, AGS6, DRADA, DSH, DSRAD, G1P1, IFI-4, IFI4, K88DSRBP, P136 | adenosine deaminase, RNA-specific (EC:3.5.4.37) |
C00044491
|
| 104 | ADARB1, ADAR2, DRABA2, DRADA2, RED1 | adenosine deaminase, RNA-specific, B1 (EC:3.5.4.37) |
C00044491
|
| 105 | ADARB2, ADAR3, RED2 | adenosine deaminase, RNA-specific, B2 (non-functional) |
C00044491
|
| 23536 | ADAT1, HADAT1 | adenosine deaminase, tRNA-specific 1 (EC:3.5.4.34) |
C00044491
|
| 90956 | ADCK2, AARF | aarF domain containing kinase 2 |
C00044491
|
| 56997 | ADCK3, ARCA2, CABC1, COQ10D4, COQ8, SCAR9 | aarF domain containing kinase 3 |
C00044491
|
| 115 | ADCY9, AC9 | adenylate cyclase 9 (EC:4.6.1.1) |
C00044491
|
| 118 | ADD1, ADDA | adducin 1 (alpha) |
C00044491
|
| 120 | ADD3, ADDL | adducin 3 (gamma) |
C00044491
|
| 126 | ADH1C, ADH3 | alcohol dehydrogenase 1C (class I), gamma polypeptide (EC:1.1.1.1) |
C00044491
|
| 128 | ADH5, ADH-3, ADHX, FALDH, FDH, GSH-FDH, GSNOR | alcohol dehydrogenase 5 (class III), chi polypeptide (EC:1.1.1.1 1.1.1.284) |
C00044491
|
| 133 | ADM, AM | adrenomedullin |
C00044491
|
| 23394 | ADNP, ADNP1 | activity-dependent neuroprotector homeobox |
C00044491
|
| 56985 | ADPRM, C17orf48, NBLA03831 | ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent (EC:3.6.1.13 3.6.1.16 3.6.1.53) |
C00044491
|
| 150 | ADRA2A, ADRA2, ADRA2R, ADRAR, ALPHA2AAR, ZNF32 | adrenoceptor alpha 2A |
C00044491
|
| 151 | ADRA2B, ADRA2L1, ADRA2RL1, ADRARL1, ALPHA2BAR, alpha-2BAR | adrenoceptor alpha 2B |
C00044491
|
| 158 | ADSL, AMPS, ASASE, ASL | adenylosuccinate lyase (EC:4.3.2.2) |
C00044491
|
| 27125 | AFF4, AF5Q31, MCEF | AF4/FMR2 family, member 4 |
C00044491
|
| 174 | AFP, FETA, HPAFP | alpha-fetoprotein |
C00044491
|
| 116987 | AGAP1, AGAP-1, CENTG2, GGAP1, cnt-g2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
C00044491
|
| 79841 | AGBL2 | ATP/GTP binding protein-like 2 |
C00044491
|
| 60509 | AGBL5 | ATP/GTP binding protein-like 5 |
C00044491
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00044491
|
| 3267 | AGFG1, HRB, RAB, RIP | ArfGAP with FG repeats 1 |
C00044491
|
| 3268 | AGFG2, HRBL, RABR | ArfGAP with FG repeats 2 |
C00044491
|
| 55109 | AGGF1, GPATC7, GPATCH7, HSU84971, HUS84971, VG5Q | angiogenic factor with G patch and FHA domains 1 |
C00044491
|
| 178 | AGL, GDE | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase (EC:2.4.1.25 3.2.1.33) |
C00044491
|
| 192670 | AGO4, EIF2C4 | argonaute RISC catalytic component 4 |
C00044491
|
| 10555 | AGPAT2, 1-AGPAT2, BSCL, BSCL1, LPAAB, LPAAT-beta | 1-acylglycerol-3-phosphate O-acyltransferase 2 (EC:2.3.1.51) |
C00044491
|
| 56894 | AGPAT3, LPAAT-GAMMA1, LPAAT3 | 1-acylglycerol-3-phosphate O-acyltransferase 3 (EC:2.3.1.51) |
C00044491
|
| 137964 | AGPAT6, 1-AGPAT_6, LPAAT-zeta, LPAATZ, TSARG7 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (EC:2.3.1.15) |
C00044491
|
| 84803 | AGPAT9, AGPAT_10, AGPAT8, GPAT3, LPAAT-theta, MAG1 | 1-acylglycerol-3-phosphate O-acyltransferase 9 (EC:2.3.1.51 2.3.1.15) |
C00044491
|
| 8540 | AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY | alkylglycerone phosphate synthase (EC:2.5.1.26) |
C00044491
|
| 23287 | AGTPBP1, CCP1, NNA1 | ATP/GTP binding protein 1 |
C00044491
|
| 185 | AGTR1, AG2S, AGTR1A, AGTR1B, AT1, AT1AR, AT1B, AT1BR, AT1R, AT2R1, AT2R1A, AT2R1B, HAT1R | angiotensin II receptor, type 1 |
C00044491
|
| 57085 | AGTRAP, ATRAP | angiotensin II receptor-associated protein |
C00044491
|
| 64902 | AGXT2, AGT2, DAIBAT | alanine--glyoxylate aminotransferase 2 (EC:2.6.1.44 2.6.1.40) |
C00044491
|
| 25909 | AHCTF1, ELYS, MST108, TMBS62 | AT hook containing transcription factor 1 |
C00044491
|
| 191 | AHCY, SAHH, adoHcyase | adenosylhomocysteinase (EC:3.3.1.1) |
C00044491
|
| 27245 | AHDC1, CL23945, DJ159A19.3, RP1-159A19.1 | AT hook, DNA binding motif, containing 1 |
C00044491
|
| 79026 | AHNAK, AHNAKRS | AHNAK nucleoprotein |
C00044491
|
| 197 | AHSG, A2HS, AHS, FETUA, HSGA | alpha-2-HS-glycoprotein |
C00044491
|
| 64853 | AIDA, C1orf80, RP11-378J18.7 | axin interactor, dorsalization associated |
C00044491
|
| 202 | AIM1, CRYBG1, ST4 | absent in melanoma 1 |
C00044491
|
| 326 | AIRE, AIRE1, APECED, APS1, APSI, PGA1 | autoimmune regulator |
C00044491
|
| 84962 | AJUBA, JUB | ajuba LIM protein |
C00044491
|
| 203 | AK1 | adenylate kinase 1 (EC:2.7.4.3 2.7.4.6) |
C00044491
|
| 204 | AK2, ADK2, AK_2 | adenylate kinase 2 (EC:2.7.4.3) |
C00044491
|
| 11216 | AKAP10, AKAP-10, D-AKAP-2, D-AKAP2, PRKA10 | A kinase (PRKA) anchor protein 10 |
C00044491
|
| 11215 | AKAP11, AKAP-11, AKAP220, PPP1R44, PRKA11 | A kinase (PRKA) anchor protein 11 |
C00044491
|
| 9590 | AKAP12, AKAP250, SSeCKS | A kinase (PRKA) anchor protein 12 |
C00044491
|
| 9465 | AKAP7, AKAP15, AKAP18 | A kinase (PRKA) anchor protein 7 |
C00044491
|
| 10142 | AKAP9, AKAP-9, AKAP350, AKAP450, CG-NAP, HYPERION, LQT11, MU-RMS-40.16A, PPP1R45, PRKA9, YOTIAO | A kinase (PRKA) anchor protein 9 |
C00044491
|
| 56672 | AKIP1, BCA3, C11orf17 | A kinase (PRKA) interacting protein 1 |
C00044491
|
| 79647 | AKIRIN1, C1orf108, RP11-781D11.2, STRF2 | akirin 1 |
C00044491
|
| 55122 | AKIRIN2, C6orf166, FBI1, dJ486L4.2 | akirin 2 |
C00044491
|
| 231 | AKR1B1, ADR, ALDR1, ALR2, AR | aldo-keto reductase family 1, member B1 (aldose reductase) (EC:1.1.1.21) |
C00044491
|
| 57016 | AKR1B10, AKR1B11, AKR1B12, ALDRLn, ARL-1, ARL1, HIS, HSI | aldo-keto reductase family 1, member B10 (aldose reductase) (EC:1.1.1.2) |
C00044491
|
| 1645 | AKR1C1, 2-ALPHA-HSD, 20-ALPHA-HSD, C9, DD1, DD1/DD2, DDH, DDH1, H-37, HAKRC, HBAB, MBAB | aldo-keto reductase family 1, member C1 (EC:1.3.1.20 1.1.1.149 1.1.1.112) |
C00044491
|
| 8644 | AKR1C3, DD3, DDX, HA1753, HAKRB, HAKRe, HSD17B5, PGFS, hluPGFS | aldo-keto reductase family 1, member C3 (EC:1.3.1.20 1.1.1.188 1.1.1.239 1.1.1.64 1.1.1.112 1.1.1.357) |
C00044491
|
| 211 | ALAS1, ALAS, ALAS3, ALASH, MIG4 | aminolevulinate, delta-, synthase 1 (EC:2.3.1.37) |
C00044491
|
| 213 | ALB, PRO0883, PRO0903, PRO1341 | albumin |
C00044491
|
| 214 | ALCAM, CD166, MEMD | activated leukocyte cell adhesion molecule |
C00044491
|
| 5832 | ALDH18A1, ARCL3A, GSAS, P5CS, PYCS | aldehyde dehydrogenase 18 family, member A1 (EC:1.2.1.41 2.7.2.11) |
C00044491
|
| 216 | ALDH1A1, ALDC, ALDH-E1, ALDH1, ALDH11, PUMB1, RALDH1 | aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) |
C00044491
|
| 224 | ALDH3A2, ALDH10, FALDH, SLS | aldehyde dehydrogenase 3 family, member A2 (EC:1.2.1.3) |
C00044491
|
| 64577 | ALDH8A1, ALDH12, DJ352A20.2 | aldehyde dehydrogenase 8 family, member A1 |
C00044491
|
| 199857 | ALG14 | ALG14, UDP-N-acetylglucosaminyltransferase subunit |
C00044491
|
| 249 | ALPL, AP-TNAP, APTNAP, HOPS, TNAP, TNSALP | alkaline phosphatase, liver/bone/kidney (EC:3.1.3.1) |
C00044491
|
| 57679 | ALS2, ALS2CR6, ALSJ, IAHSP, PLSJ | amyotrophic lateral sclerosis 2 (juvenile) |
C00044491
|
| 262 | AMD1, ADOMETDC, AMD, SAMDC | adenosylmethionine decarboxylase 1 (EC:4.1.1.50) |
C00044491
|
| 267 | AMFR, GP78, RNF45 | autocrine motility factor receptor, E3 ubiquitin protein ligase (EC:6.3.2.19) |
C00044491
|
| 268 | AMH, MIF, MIS | anti-Mullerian hormone |
C00044491
|
| 347902 | AMIGO2, ALI1, AMIGO-2, DEGA | adhesion molecule with Ig-like domain 2 |
C00044491
|
| 9949 | AMMECR1, AMMERC1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
C00044491
|
| 83607 | AMMECR1L | AMMECR1-like |
C00044491
|
| 154810 | AMOTL1, JEAP | angiomotin like 1 |
C00044491
|
| 51421 | AMOTL2, LCCP | angiomotin like 2 |
C00044491
|
| 271 | AMPD2 | adenosine monophosphate deaminase 2 (EC:3.5.4.6) |
C00044491
|
| 272 | AMPD3 | adenosine monophosphate deaminase 3 (EC:3.5.4.6) |
C00044491
|
| 51321 | AMZ2 | archaelysin family metallopeptidase 2 |
C00044491
|
| 119504 | ANAPC16, APC16, C10orf104, CENP-27, MSAG, bA570G20.3 | anaphase promoting complex subunit 16 |
C00044491
|
| 51433 | ANAPC5, APC5 | anaphase promoting complex subunit 5 |
C00044491
|
| 283 | ANG, ALS9, HEL168, RNASE4, RNASE5 | angiogenin, ribonuclease, RNase A family, 5 (EC:3.1.27.-) |
C00044491
|
| 23452 | ANGPTL2, ARP2, HARP | angiopoietin-like 2 |
C00044491
|
| 51479 | ANKFY1, ANKHZN, BTBD23, ZFYVE14 | ankyrin repeat and FYVE domain containing 1 |
C00044491
|
| 56172 | ANKH, ANK, CCAL2, CMDJ, CPPDD, HANK, MANK | ANKH inorganic pyrophosphate transport regulator |
C00044491
|
| 54467 | ANKIB1 | ankyrin repeat and IBR domain containing 1 |
C00044491
|
| 57037 | ANKMY2, ZMYND20 | ankyrin repeat and MYND domain containing 2 |
C00044491
|
| 29123 | ANKRD11, ANCO-1, ANCO1, LZ16, T13 | ankyrin repeat domain 11 |
C00044491
|
| 23253 | ANKRD12, ANCO-2, ANCO1, GAC-1, Nbla00144 | ankyrin repeat domain 12 |
C00044491
|
| 88455 | ANKRD13A, ANKRD13, NY-REN-25 | ankyrin repeat domain 13A |
C00044491
|
| 54522 | ANKRD16 | ankyrin repeat domain 16 |
C00044491
|
| 26057 | ANKRD17, GTAR, NY-BR-16 | ankyrin repeat domain 17 |
C00044491
|
| 22852 | ANKRD26, THC2, bA145E8.1 | ankyrin repeat domain 26 |
C00044491
|
| 23243 | ANKRD28, PITK, PPP1R65 | ankyrin repeat domain 28 |
C00044491
|
| 84250 | ANKRD32, BRCTD1, BRCTx | ankyrin repeat domain 32 |
C00044491
|
| 651746 | ANKRD33B | ankyrin repeat domain 33B |
C00044491
|
| 57730 | ANKRD36B, KIAA1641 | ankyrin repeat domain 36B |
C00044491
|
| 353322 | ANKRD37, Lrp2bp | ankyrin repeat domain 37 |
C00044491
|
| 23294 | ANKS1A, ANKS1 | ankyrin repeat and sterile alpha motif domain containing 1A |
C00044491
|
| 203286 | ANKS6, ANKRD14, NPHP16, PKDR1, SAMD6 | ankyrin repeat and sterile alpha motif domain containing 6 |
C00044491
|
| 54443 | ANLN, Scraps, scra | anillin, actin binding protein |
C00044491
|
| 8125 | ANP32A, C15orf1, HPPCn, I1PP2A, LANP, MAPM, PHAP1, PHAPI, PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
C00044491
|
| 81611 | ANP32E, LANP-L, LANPL | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
C00044491
|
| 118429 | ANTXR2, CMG-2, CMG2, HFS, ISH, JHF | anthrax toxin receptor 2 |
C00044491
|
| 11199 | ANXA10, ANX14 | annexin A10 |
C00044491
|
| 311 | ANXA11, ANX11, CAP50 | annexin A11 |
C00044491
|
| 307 | ANXA4, ANX4, PIG28, ZAP36 | annexin A4 |
C00044491
|
| 310 | ANXA7, ANX7, SNX, SYNEXIN | annexin A7 |
C00044491
|
| 164 | AP1G1, ADTG, CLAPG1 | adaptor-related protein complex 1, gamma 1 subunit |
C00044491
|
| 8906 | AP1G2, G2AD | adaptor-related protein complex 1, gamma 2 subunit |
C00044491
|
| 8907 | AP1M1, AP47, CLAPM2, CLTNM, MU-1A | adaptor-related protein complex 1, mu 1 subunit |
C00044491
|
| 1174 | AP1S1, AP19, CLAPS1, MEDNIK, SIGMA1A, WUGSC:H_DJ0747G18.2 | adaptor-related protein complex 1, sigma 1 subunit |
C00044491
|
| 8905 | AP1S2, MGC:1902, MRX59, MRXS21, MRXSF, SIGMA1B | adaptor-related protein complex 1, sigma 2 subunit |
C00044491
|
| 130340 | AP1S3 | adaptor-related protein complex 1, sigma 3 subunit |
C00044491
|
| 160 | AP2A1, ADTAA, AP2-ALPHA, CLAPA1 | adaptor-related protein complex 2, alpha 1 subunit |
C00044491
|
| 8943 | AP3D1, ADTD, hBLVR | adaptor-related protein complex 3, delta 1 subunit |
C00044491
|
| 55745 | AP5M1, C14orf108, MUDENG, Mu5, MuD | adaptor-related protein complex 5, mu 1 subunit |
C00044491
|
| 317 | APAF1, APAF-1, CED4 | apoptotic peptidase activating factor 1 |
C00044491
|
| 54518 | APBB1IP, INAG1, PREL1, RARP1, RIAM | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
C00044491
|
| 323 | APBB2, FE65L, FE65L1 | amyloid beta (A4) precursor protein-binding, family B, member 2 |
C00044491
|
| 327 | APEH, AARE, ACPH, APH, D3F15S2, D3S48E, DNF15S2, OPH | acylaminoacyl-peptide hydrolase (EC:3.4.19.1) |
C00044491
|
| 27301 | APEX2, APE2, APEXL2, XTH2 | APEX nuclease (apurinic/apyrimidinic endonuclease) 2 (EC:4.2.99.18) |
C00044491
|
| 333 | APLP1, APLP | amyloid beta (A4) precursor-like protein 1 |
C00044491
|
| 334 | APLP2, APLP-2, APPH, APPL2, CDEBP | amyloid beta (A4) precursor-like protein 2 |
C00044491
|
| 57136 | APMAP, BSCv, C20orf3 | adipocyte plasma membrane associated protein |
C00044491
|
| 128240 | APOA1BP, AIBP, YJEFN1 | apolipoprotein A-I binding protein (EC:5.1.99.6) |
C00044491
|
| 337 | APOA4 | apolipoprotein A-IV |
C00044491
|
| 338 | APOB, FLDB, LDLCQ4 | apolipoprotein B |
C00044491
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00044491
|
| 27350 | APOBEC3C, A3C, APOBEC1L, ARDC2, ARDC4, ARP5, PBI, bK150C2.3 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3C |
C00044491
|
| 319 | APOF, Apo-F, LTIP | apolipoprotein F |
C00044491
|
| 350 | APOH, B2G1, B2GP1, BG | apolipoprotein H (beta-2-glycoprotein I) |
C00044491
|
| 8542 | APOL1, APO-L, APOL, APOL-I, FSGS4 | apolipoprotein L, 1 |
C00044491
|
| 23780 | APOL2, APOL-II, APOL3 | apolipoprotein L, 2 |
C00044491
|
| 80833 | APOL3, APOLIII, CG121 | apolipoprotein L, 3 |
C00044491
|
| 80830 | APOL6, APOL-VI, APOLVI | apolipoprotein L, 6 |
C00044491
|
| 81575 | APOLD1, VERGE | apolipoprotein L domain containing 1 |
C00044491
|
| 139322 | APOOL, CXorf33, FAM121A, UNQ8193 | apolipoprotein O-like |
C00044491
|
| 10513 | APPBP2, APP-BP2, HS.84084, PAT1 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
C00044491
|
| 360 | AQP3, AQP-3, GIL | aquaporin 3 (Gill blood group) |
C00044491
|
| 364 | AQP7, AQP7L, AQP9, AQPap, GLYCQTL | aquaporin 7 |
C00044491
|
| 366 | AQP9, AQP-9, HsT17287, SSC1 | aquaporin 9 |
C00044491
|
| 9716 | AQR, IBP160, fSAP164 | aquarius intron-binding spliceosomal factor |
C00044491
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00044491
|
| 374 | AREG, AR, CRDGF, SDGF | amphiregulin |
C00044491
|
| 9870 | AREL1, KIAA0317 | apoptosis resistant E3 ubiquitin protein ligase 1 |
C00044491
|
| 377 | ARF3 | ADP-ribosylation factor 3 |
C00044491
|
| 10565 | ARFGEF1, ARFGEP1, BIG1, P200 | ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
C00044491
|
| 10564 | ARFGEF2, BIG2, PVNH2, dJ1164I10.1 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
C00044491
|
| 384 | ARG2 | arginase 2 (EC:3.5.3.1) |
C00044491
|
| 392 | ARHGAP1, CDC42GAP, RHOGAP, RHOGAP1, p50rhoGAP | Rho GTPase activating protein 1 |
C00044491
|
| 9824 | ARHGAP11A, GAP_(1-12) | Rho GTPase activating protein 11A |
C00044491
|
| 89839 | ARHGAP11B, B'-T, FAM7B1 | Rho GTPase activating protein 11B |
C00044491
|
| 94134 | ARHGAP12 | Rho GTPase activating protein 12 |
C00044491
|
| 93663 | ARHGAP18, MacGAP, SENEX, bA307O14.2 | Rho GTPase activating protein 18 |
C00044491
|
| 84986 | ARHGAP19 | Rho GTPase activating protein 19 |
C00044491
|
| 57569 | ARHGAP20, RARHOGAP | Rho GTPase activating protein 20 |
C00044491
|
| 23092 | ARHGAP26, GRAF, GRAF1, OPHN1L, OPHN1L1 | Rho GTPase activating protein 26 |
C00044491
|
| 2909 | ARHGAP35, GRF-1, GRLF1, P190-A, P190A, p190ARhoGAP, p190RhoGAP | Rho GTPase activating protein 35 |
C00044491
|
| 396 | ARHGDIA, GDIA1, NPHS8, RHOGDI, RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha |
C00044491
|
| 397 | ARHGDIB, D4, GDIA2, GDID4, LYGDI, Ly-GDI, RAP1GN1, RhoGDI2 | Rho GDP dissociation inhibitor (GDI) beta |
C00044491
|
| 9639 | ARHGEF10, GEF10 | Rho guanine nucleotide exchange factor (GEF) 10 |
C00044491
|
| 55160 | ARHGEF10L, GrinchGEF | Rho guanine nucleotide exchange factor (GEF) 10-like |
C00044491
|
| 23365 | ARHGEF12, LARG, PRO2792 | Rho guanine nucleotide exchange factor (GEF) 12 |
C00044491
|
| 50650 | ARHGEF3, GEF3, STA3, XPLN | Rho guanine nucleotide exchange factor (GEF) 3 |
C00044491
|
| 8874 | ARHGEF7, BETA-PIX, COOL-1, COOL1, Nbla10314, P50, P50BP, P85, P85COOL1, P85SPR, PAK3, PIXB | Rho guanine nucleotide exchange factor (GEF) 7 |
C00044491
|
| 8289 | ARID1A, B120, BAF250, BAF250a, BM029, C1orf4, ELD, MRD14, OSA1, P270, SMARCF1, hELD, hOSA1 | AT rich interactive domain 1A (SWI-like) |
C00044491
|
| 1820 | ARID3A, BRIGHT, DRIL1, DRIL3, E2FBP1 | AT rich interactive domain 3A (BRIGHT-like) |
C00044491
|
| 5926 | ARID4A, RBBP-1, RBBP1, RBP-1, RBP1 | AT rich interactive domain 4A (RBP1-like) |
C00044491
|
| 25820 | ARIH1, ARI, HARI, HHARI, UBCH7BP | ariadne RBR E3 ubiquitin protein ligase 1 |
C00044491
|
| 80117 | ARL14, ARF7 | ADP-ribosylation factor-like 14 |
C00044491
|
| 54622 | ARL15, ARFRP2 | ADP-ribosylation factor-like 15 |
C00044491
|
| 403 | ARL3, ARFL3 | ADP-ribosylation factor-like 3 |
C00044491
|
| 379 | ARL4D, ARF4L, ARL6 | ADP-ribosylation factor-like 4D |
C00044491
|
| 23204 | ARL6IP1, AIP1, ARL6IP, ARMER | ADP-ribosylation factor-like 6 interacting protein 1 |
C00044491
|
| 51329 | ARL6IP4, SFRS20, SR-25, SRp25, SRrp37 | ADP-ribosylation-like factor 6 interacting protein 4 |
C00044491
|
| 10550 | ARL6IP5, DERP11, GTRAP3-18, JWA, PRAF3, addicsin, hp22, jmx | ADP-ribosylation-like factor 6 interacting protein 5 |
C00044491
|
| 151188 | ARL6IP6, AIP-6, PFAAP1 | ADP-ribosylation-like factor 6 interacting protein 6 |
C00044491
|
| 127829 | ARL8A, ARL10B, GIE2 | ADP-ribosylation factor-like 8A |
C00044491
|
| 51309 | ARMCX1, ALEX1 | armadillo repeat containing, X-linked 1 |
C00044491
|
| 64860 | ARMCX5 | armadillo repeat containing, X-linked 5 |
C00044491
|
| 56938 | ARNTL2, BMAL2, CLIF, MOP9, PASD9, bHLHe6 | aryl hydrocarbon receptor nuclear translocator-like 2 |
C00044491
|
| 10552 | ARPC1A, Arc40, SOP2Hs, SOP2L | actin related protein 2/3 complex, subunit 1A, 41kDa |
C00044491
|
| 10776 | ARPP19, ARPP-16, ARPP-19, ARPP16, ENSAL | cAMP-regulated phosphoprotein, 19kDa |
C00044491
|
| 407 | ARR3, ARRX | arrestin 3, retinal (X-arrestin) |
C00044491
|
| 408 | ARRB1, ARB1, ARR1 | arrestin, beta 1 |
C00044491
|
| 409 | ARRB2, ARB2, ARR2, BARR2 | arrestin, beta 2 |
C00044491
|
| 57561 | ARRDC3, TLIMP | arrestin domain containing 3 |
C00044491
|
| 91947 | ARRDC4 | arrestin domain containing 4 |
C00044491
|
| 153642 | ARSK, TSULF | arylsulfatase family, member K |
C00044491
|
| 420 | ART4, ARTC4, CD297, DO, DOK1 | ADP-ribosyltransferase 4 (EC:2.4.2.31) |
C00044491
|
| 421 | ARVCF | armadillo repeat gene deleted in velocardiofacial syndrome |
C00044491
|
| 57412 | AS3MT, CYT19 | arsenic (+3 oxidation state) methyltransferase (EC:2.1.1.137) |
C00044491
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00044491
|
| 55616 | ASAP3, ACAP4, CENTB6, DDEFL1, UPLC1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
C00044491
|
| 339201 | ASB16-AS1, C17orf65 | ASB16 antisense RNA 1 |
C00044491
|
| 140460 | ASB7 | ankyrin repeat and SOCS box containing 7 |
C00044491
|
| 10973 | ASCC3, ASC1p200, HELIC1, RNAH | activating signal cointegrator 1 complex subunit 3 (EC:3.6.4.12) |
C00044491
|
| 121549 | ASCL4, HASH4, bHLHa44 | achaete-scute family bHLH transcription factor 4 |
C00044491
|
| 55723 | ASF1B, CIA-II | anti-silencing function 1B histone chaperone |
C00044491
|
| 432 | ASGR1, ASGPR, ASGPR1, CLEC4H1, HL-1 | asialoglycoprotein receptor 1 |
C00044491
|
| 440 | ASNS, TS11 | asparagine synthetase (glutamine-hydrolyzing) (EC:6.3.5.4) |
C00044491
|
| 374569 | ASPG, C14orf76, GPA/WT | asparaginase homolog (S. cerevisiae) (EC:3.1.1.5 3.1.1.47 3.5.1.1) |
C00044491
|
| 444 | ASPH, AAH, BAH, CASQ2BP1, HAAH, JCTN, junctin | aspartate beta-hydroxylase (EC:1.14.11.16) |
C00044491
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00044491
|
| 460 | ASTN1, ASTN | astrotactin 1 |
C00044491
|
| 55726 | ASUN, C12orf11, GCT1, Mat89Bb, NET48, SPATA30 | asunder spermatogenesis regulator |
C00044491
|
| 171023 | ASXL1, BOPS, MDS | additional sex combs like 1 (Drosophila) |
C00044491
|
| 29028 | ATAD2, ANCCA, PRO2000 | ATPase family, AAA domain containing 2 (EC:3.6.1.3) |
C00044491
|
| 79915 | ATAD5, C17orf41, ELG1, FRAG1 | ATPase family, AAA domain containing 5 |
C00044491
|
| 11101 | ATE1 | arginyltransferase 1 (EC:2.3.2.8) |
C00044491
|
| 466 | ATF1, EWS-ATF1, FUS/ATF-1, TREB36 | activating transcription factor 1 |
C00044491
|
| 467 | ATF3 | activating transcription factor 3 |
C00044491
|
| 468 | ATF4, CREB-2, CREB2, TAXREB67, TXREB | activating transcription factor 4 |
C00044491
|
| 55729 | ATF7IP, AM, ATF-IP, MCAF, MCAF1, p621 | activating transcription factor 7 interacting protein |
C00044491
|
| 80063 | ATF7IP2, MCAF2 | activating transcription factor 7 interacting protein 2 |
C00044491
|
| 9140 | ATG12, APG12, APG12L, FBR93, HAPG12 | autophagy related 12 |
C00044491
|
| 55054 | ATG16L1, APG16L, ATG16A, ATG16L, IBD10, WDR30 | autophagy related 16-like 1 (S. cerevisiae) |
C00044491
|
| 89849 | ATG16L2, ATG16B, WDR80 | autophagy related 16-like 2 (S. cerevisiae) |
C00044491
|
| 55102 | ATG2B, C14orf103 | autophagy related 2B |
C00044491
|
| 115201 | ATG4A, APG4A, AUTL2 | autophagy related 4A, cysteine peptidase |
C00044491
|
| 471 | ATIC, AICAR, AICARFT, IMPCHASE, PURH | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (EC:2.1.2.3 3.5.4.10) |
C00044491
|
| 25923 | ATL3 | atlastin GTPase 3 |
C00044491
|
| 472 | ATM, AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | ataxia telangiectasia mutated (EC:2.7.11.1) |
C00044491
|
| 474 | ATOH1, ATH1, HATH1, MATH-1, bHLHa14 | atonal homolog 1 (Drosophila) |
C00044491
|
| 57205 | ATP10D, ATPVD | ATPase, class V, type 10D (EC:3.6.3.1) |
C00044491
|
| 23250 | ATP11A, ATPIH, ATPIS | ATPase, class VI, type 11A (EC:3.6.3.1) |
C00044491
|
| 23200 | ATP11B, ATPIF, ATPIR | ATPase, class VI, type 11B (EC:3.6.3.1) |
C00044491
|
| 286410 | ATP11C, ATPIG, ATPIQ | ATPase, class VI, type 11C (EC:3.6.3.1) |
C00044491
|
| 57130 | ATP13A1, ATP13A | ATPase type 13A1 (EC:3.6.3.8) |
C00044491
|
| 79572 | ATP13A3, AFURS1 | ATPase type 13A3 |
C00044491
|
| 476 | ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (EC:3.6.3.9) |
C00044491
|
| 481 | ATP1B1, ATP1B | ATPase, Na+/K+ transporting, beta 1 polypeptide |
C00044491
|
| 488 | ATP2A2, ATP2B, DAR, DD, SERCA2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (EC:3.6.3.8) |
C00044491
|
| 490 | ATP2B1, PMCA1, PMCA1kb | ATPase, Ca++ transporting, plasma membrane 1 (EC:3.6.3.8) |
C00044491
|
| 491 | ATP2B2, PMCA2, PMCA2a, PMCA2i | ATPase, Ca++ transporting, plasma membrane 2 (EC:3.6.3.8) |
C00044491
|
| 513 | ATP5D | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit (EC:3.6.1.14) |
C00044491
|
| 515 | ATP5F1, PIG47 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
C00044491
|
| 9551 | ATP5J2, ATP5JL | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 (EC:3.6.1.14) |
C00044491
|
| 27109 | ATP5S, ATPW, FB, HSU79253 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) (EC:3.6.1.14) |
C00044491
|
| 55101 | ATP5SL | ATP5S-like |
C00044491
|
| 10159 | ATP6AP2, APT6M8-9, ATP6IP2, ATP6M8-9, ELDF10, M8-9, MRXE, MSTP009, XMRE | ATPase, H+ transporting, lysosomal accessory protein 2 |
C00044491
|
| 535 | ATP6V0A1, ATP6N1, ATP6N1A, Stv1, VPP1, Vph1, a1 | ATPase, H+ transporting, lysosomal V0 subunit a1 (EC:3.6.3.14) |
C00044491
|
| 51382 | ATP6V1D, ATP6M, VATD, VMA8 | ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D (EC:3.6.3.14) |
C00044491
|
| 5205 | ATP8B1, ATPIC, BRIC, FIC1, ICP1, PFIC, PFIC1 | ATPase, aminophospholipid transporter, class I, type 8B, member 1 (EC:3.6.3.1) |
C00044491
|
| 57198 | ATP8B2, ATPID | ATPase, aminophospholipid transporter, class I, type 8B, member 2 (EC:3.6.3.1) |
C00044491
|
| 545 | ATR, FCTCS, FRP1, MEC1, SCKL, SCKL1 | ataxia telangiectasia and Rad3 related (EC:2.7.11.1) |
C00044491
|
| 8455 | ATRN, DPPT-L, MGCA | attractin |
C00044491
|
| 546 | ATRX, ATR2, JMS, MRXHF1, RAD54, RAD54L, SFM1, SHS, XH2, XNP, ZNF-HX | alpha thalassemia/mental retardation syndrome X-linked (EC:3.6.4.12) |
C00044491
|
| 6310 | ATXN1, ATX1, D6S504E, SCA1 | ataxin 1 |
C00044491
|
| 6314 | ATXN7, ADCAII, OPCA3, SCA7 | ataxin 7 |
C00044491
|
| 222255 | ATXN7L1, ATXN7L4 | ataxin 7-like 1 |
C00044491
|
| 79000 | AUNIP, AIBP, C1orf135 | aurora kinase A and ninein interacting protein |
C00044491
|
| 60370 | AVPI1, PP5395, RP11-548K23.7, VIP32, VIT32 | arginine vasopressin-induced 1 |
C00044491
|
| 558 | AXL, JTK11, UFO | AXL receptor tyrosine kinase (EC:2.7.10.1) |
C00044491
|
| 566 | AZU1, AZAMP, AZU, CAP37, HBP, HUMAZUR, NAZC | azurocidin 1 |
C00044491
|
| 567 | B2M | beta-2-microglobulin |
C00044491
|
| 26229 | B3GAT3, GLCATI | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (EC:2.4.1.135) |
C00044491
|
| 11041 | B3GNT1, B3GN-T1, B3GNT6, BETA3GNTI, MDDGA13, iGAT, iGNT | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (EC:2.4.1.149) |
C00044491
|
| 10678 | B3GNT2, B3GN-T2, B3GNT, B3GNT-2, B3GNT1, BETA3GNT, BGNT2, BGnT-2 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (EC:2.4.1.149) |
C00044491
|
| 2683 | B4GALT1, B4GAL-T1, CDG2D, GGTB2, GT1, GTB, beta4Gal-T1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (EC:2.4.1.22 2.4.1.90 2.4.1.38) |
C00044491
|
| 9334 | B4GALT5, B4Gal-T5, BETA4-GALT-IV, beta4Gal-T5, beta4GalT-V, gt-V | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (EC:2.4.1.38) |
C00044491
|
| 9331 | B4GALT6, B4Gal-T6, beta4Gal-T6 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (EC:2.4.1.274) |
C00044491
|
| 23621 | BACE1, ASP2, BACE, HSPC104 | beta-site APP-cleaving enzyme 1 (EC:3.4.23.46) |
C00044491
|
| 25825 | BACE2, AEPLC, ALP56, ASP1, ASP21, BAE2, CEAP1, DRAP | beta-site APP-cleaving enzyme 2 (EC:3.4.23.45) |
C00044491
|
| 60468 | BACH2, BTBD25 | BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
C00044491
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00044491
|
| 7917 | BAG6, BAG-6, BAT3, D6S52E, G3 | BCL2-associated athanogene 6 |
C00044491
|
| 10458 | BAIAP2, BAP2, FLAF3, IRSP53 | BAI1-associated protein 2 |
C00044491
|
| 55971 | BAIAP2L1, IRTKS | BAI1-associated protein 2-like 1 |
C00044491
|
| 80115 | BAIAP2L2 | BAI1-associated protein 2-like 2 |
C00044491
|
| 25805 | BAMBI, NMA | BMP and activin membrane-bound inhibitor |
C00044491
|
| 10409 | BASP1, CAP-23, CAP23, NAP-22, NAP22 | brain abundant, membrane attached signal protein 1 |
C00044491
|
| 393917 |
C00044491
|
||
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00044491
|
| 92482 | BBIP1, BBIP10, NCRNA00081, bA348N5.3 | BBSome interacting protein 1 |
C00044491
|
| 583 | BBS2, BBS | Bardet-Biedl syndrome 2 |
C00044491
|
| 55212 | BBS7, BBS2L1 | Bardet-Biedl syndrome 7 |
C00044491
|
| 56987 | BBX, ARTC1, HBP2, HSPC339, MDS001 | bobby sox homolog (Drosophila) |
C00044491
|
| 10286 | BCAS2, DAM1, SPF27, Snt309 | breast carcinoma amplified sequence 2 |
C00044491
|
| 586 | BCAT1, BCATC, BCT1, ECA39, MECA39, PNAS121, PP18 | branched chain amino-acid transaminase 1, cytosolic (EC:2.6.1.42) |
C00044491
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00044491
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00044491
|
| 10018 | BCL2L11, BAM, BIM, BOD | BCL2-like 11 (apoptosis facilitator) |
C00044491
|
| 602 | BCL3, BCL4, D19S37 | B-cell CLL/lymphoma 3 |
C00044491
|
| 9275 | BCL7B | B-cell CLL/lymphoma 7B |
C00044491
|
| 9774 | BCLAF1, BTF, bK211L9.1 | BCL2-associated transcription factor 1 |
C00044491
|
| 53630 | BCMO1, BCDO, BCDO1, BCMO, BCO, BCO1 | beta-carotene 15,15'-monooxygenase 1 (EC:1.14.99.36) |
C00044491
|
| 57673 | BEND3, KIAA1553, RP11-59I9.2 | BEN domain containing 3 |
C00044491
|
| 10282 | BET1, HBET1 | Bet1 golgi vesicular membrane trafficking protein |
C00044491
|
| 84707 | BEX2, BEX1, DJ79P11.1 | brain expressed X-linked 2 |
C00044491
|
| 633 | BGN, DSPG1, PG-S1, PGI, SLRR1A | biglycan |
C00044491
|
| 23299 | BICD2, SMALED2, bA526D8.1 | bicaudal D homolog 2 (Drosophila) |
C00044491
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00044491
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00044491
|
| 57448 | BIRC6, APOLLON, BRUCE | baculoviral IAP repeat containing 6 |
C00044491
|
| 54841 | BIVM | basic, immunoglobulin-like variable motif containing |
C00044491
|
| 640 | BLK, MODY11 | B lymphoid tyrosine kinase (EC:2.7.10.2) |
C00044491
|
| 641 | BLM, BS, RECQ2, RECQL2, RECQL3 | Bloom syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00044491
|
| 29760 | BLNK, AGM4, BASH, BLNK-S, LY57, SLP-65, SLP65, bca | B-cell linker |
C00044491
|
| 282991 | BLOC1S2, BLOS2, CEAP, CEAP11, RP11-316M21.4 | biogenesis of lysosomal organelles complex-1, subunit 2 |
C00044491
|
| 388552 | BLOC1S3, BLOS3, HPS8, RP | biogenesis of lysosomal organelles complex-1, subunit 3 |
C00044491
|
| 63915 | BLOC1S5, BLOS5, MU, MUTED | biogenesis of lysosomal organelles complex-1, subunit 5, muted |
C00044491
|
| 644 | BLVRA, BLVR, BVR, BVRA | biliverdin reductase A (EC:1.3.1.24) |
C00044491
|
| 648 | BMI1, FLVI2/BMI1, PCGF4, RNF51 | BMI1 polycomb ring finger oncogene |
C00044491
|
| 649 | BMP1, OI13, PCOLC, PCP, PCP2, TLD | bone morphogenetic protein 1 (EC:3.4.24.19) |
C00044491
|
| 654 | BMP6, VGR, VGR1 | bone morphogenetic protein 6 |
C00044491
|
| 659 | BMPR2, BMPR-II, BMPR3, BMR2, BRK-3, PPH1, T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) (EC:2.7.11.30) |
C00044491
|
| 664 | BNIP3, NIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 |
C00044491
|
| 665 | BNIP3L, BNIP3a, NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like |
C00044491
|
| 666 | BOK, BCL2L9, BOKL | BCL2-related ovarian killer |
C00044491
|
| 79866 | BORA, C13orf34 | bora, aurora kinase A activator |
C00044491
|
| 10380 | BPNT1, PIP | 3'(2'), 5'-bisphosphate nucleotidase 1 (EC:3.1.3.7) |
C00044491
|
| 2186 | BPTF, FAC1, FALZ, NURF301 | bromodomain PHD finger transcription factor |
C00044491
|
| 673 | BRAF, B-RAF1, BRAF1, NS7, RAFB1 | v-raf murine sarcoma viral oncogene homolog B (EC:2.7.11.1) |
C00044491
|
| 8315 | BRAP, BRAP2, IMP, RNF52 | BRCA1 associated protein |
C00044491
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00044491
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00044491
|
| 6046 | BRD2, D6S113E, FSH, FSRG1, NAT, RING3, RNF3 | bromodomain containing 2 |
C00044491
|
| 29117 | BRD7, BP75, CELTIX1, NAG4 | bromodomain containing 7 |
C00044491
|
| 9577 | BRE, BRCC4, BRCC45 | brain and reproductive organ-expressed (TNFRSF1A modulator) |
C00044491
|
| 140707 | BRI3BP, BNAS1, HCCR-1, HCCR-2, HCCRBP-1, KG19 | BRI3 binding protein |
C00044491
|
| 83990 | BRIP1, BACH1, FANCJ, OF | BRCA1 interacting protein C-terminal helicase 1 (EC:3.6.4.13) |
C00044491
|
| 84312 | BRMS1L, BRMS1 | breast cancer metastasis-suppressor 1-like |
C00044491
|
| 55727 | BTBD7, FUP1 | BTB (POZ) domain containing 7 |
C00044491
|
| 284697 | BTBD8 | BTB (POZ) domain containing 8 |
C00044491
|
| 114781 | BTBD9, dJ322I12.1 | BTB (POZ) domain containing 9 |
C00044491
|
| 91408 | BTF3L4 | basic transcription factor 3-like 4 |
C00044491
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00044491
|
| 10950 | BTG3, ANA, TOB5, TOB55, TOFA | BTG family, member 3 |
C00044491
|
| 11118 | BTN3A2, BT3.2, BTF4, CD277 | butyrophilin, subfamily 3, member A2 |
C00044491
|
| 10384 | BTN3A3, BTF3 | butyrophilin, subfamily 3, member A3 |
C00044491
|
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00044491
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00044491
|
| 9184 | BUB3, BUB3L, hBUB3 | BUB3 mitotic checkpoint protein |
C00044491
|
| 11067 | C10orf10, DEPP, FIG | chromosome 10 open reading frame 10 |
C00044491
|
| 26098 | C10orf137, EDRF1 | chromosome 10 open reading frame 137 |
C00044491
|
| 56652 | C10orf2, ATXN8, IOSCA, MTDPS7, PEO, PEO1, PEOA3, SANDO, SCA8, TWINL | chromosome 10 open reading frame 2 (EC:3.6.4.12) |
C00044491
|
| 64115 | C10orf54, B7-H5, B7H5, GI24, PP2135, SISP1 | chromosome 10 open reading frame 54 |
C00044491
|
| 79946 | C10orf95 | chromosome 10 open reading frame 95 |
C00044491
|
| 53838 | C11orf24, DM4E3 | chromosome 11 open reading frame 24 |
C00044491
|
| 91894 | C11orf52 | chromosome 11 open reading frame 52 |
C00044491
|
| 220042 | C11orf82, noxin | chromosome 11 open reading frame 82 |
C00044491
|
| 144097 | C11orf84 | chromosome 11 open reading frame 84 |
C00044491
|
| 65998 | C11orf95 | chromosome 11 open reading frame 95 |
C00044491
|
| 57103 | C12orf5, FR2BP, TIGAR | chromosome 12 open reading frame 5 (EC:3.1.3.46) |
C00044491
|
| 91574 | C12orf65, COXPD7, SPG55 | chromosome 12 open reading frame 65 |
C00044491
|
| 55195 | C14orf105 | chromosome 14 open reading frame 105 |
C00044491
|
| 283687 | C15orf37 | chromosome 15 open reading frame 37 |
C00044491
|
| 56905 | C15orf39 | chromosome 15 open reading frame 39 |
C00044491
|
| 730094 | C16orf52 | chromosome 16 open reading frame 52 |
C00044491
|
| 57020 | C16orf62 | chromosome 16 open reading frame 62 |
C00044491
|
| 29035 | C16orf72, PRO0149 | chromosome 16 open reading frame 72 |
C00044491
|
| 29105 | C16orf80, EVORF, GTL3, fSAP23 | chromosome 16 open reading frame 80 |
C00044491
|
| 146556 | C16orf89 | chromosome 16 open reading frame 89 |
C00044491
|
| 256302 | C17orf103, Gtlf3b | chromosome 17 open reading frame 103 |
C00044491
|
| 78995 | C17orf53 | chromosome 17 open reading frame 53 |
C00044491
|
| 64149 | C17orf75, NJMU-R1 | chromosome 17 open reading frame 75 |
C00044491
|
| 147339 | C18orf25, ARKL1 | chromosome 18 open reading frame 25 |
C00044491
|
| 162681 | C18orf54, LAS2 | chromosome 18 open reading frame 54 |
C00044491
|
| 494514 | C18orf56 | chromosome 18 open reading frame 56 |
C00044491
|
| 29919 | C18orf8, HsT2591, MIC1, Mic-1 | chromosome 18 open reading frame 8 |
C00044491
|
| 83636 | C19orf12, NBIA3, NBIA4, SPG43 | chromosome 19 open reading frame 12 |
C00044491
|
| 55009 | C19orf24 | chromosome 19 open reading frame 24 |
C00044491
|
| 91442 | C19orf40, FAAP24 | chromosome 19 open reading frame 40 |
C00044491
|
| 55337 | C19orf66 | chromosome 19 open reading frame 66 |
C00044491
|
| 29071 | C1GALT1C1, C1GALT2, C38H2-L1, COSMC, MST143, TNPS | C1GALT1-specific chaperone 1 |
C00044491
|
| 128061 | C1orf131 | chromosome 1 open reading frame 131 |
C00044491
|
| 199920 | C1orf168 | chromosome 1 open reading frame 168 |
C00044491
|
| 149466 | C1orf210 | chromosome 1 open reading frame 210 |
C00044491
|
| 79078 | C1orf50 | chromosome 1 open reading frame 50 |
C00044491
|
| 148523 | C1orf51 | chromosome 1 open reading frame 51 |
C00044491
|
| 148423 | C1orf52, RP11-234D19.1, gm117 | chromosome 1 open reading frame 52 |
C00044491
|
| 57035 | C1orf63, DJ465N24.2.1, NPD014, RP3-465N24.4 | chromosome 1 open reading frame 63 |
C00044491
|
| 140688 | C20orf112, C20orf113, dJ1184F4.2, dJ1184F4.4 | chromosome 20 open reading frame 112 |
C00044491
|
| 54058 | C21orf58 | chromosome 21 open reading frame 58 |
C00044491
|
| 79640 | C22orf46 | chromosome 22 open reading frame 46 |
C00044491
|
| 25966 | C2CD2, C21orf25, C21orf258, TMEM24L | C2 calcium-dependent domain containing 2 |
C00044491
|
| 26005 | C2CD3 | C2 calcium-dependent domain containing 3 |
C00044491
|
| 9847 | C2CD5, CDP138, KIAA0528 | C2 calcium-dependent domain containing 5 |
C00044491
|
| 79568 | C2orf47 | chromosome 2 open reading frame 47 |
C00044491
|
| 79919 | C2orf54 | chromosome 2 open reading frame 54 |
C00044491
|
| 257407 | C2orf72 | chromosome 2 open reading frame 72 |
C00044491
|
| 130355 | C2orf76, AIM29 | chromosome 2 open reading frame 76 |
C00044491
|
| 389084 | C2orf82, UNQ830 | chromosome 2 open reading frame 82 |
C00044491
|
| 719 | C3AR1, AZ3B, C3AR, HNFAG09 | complement component 3a receptor 1 |
C00044491
|
| 205428 | C3orf58, DIA1, GoPro49, HASF | chromosome 3 open reading frame 58 |
C00044491
|
| 388503 | C3P1, CPLP | complement component 3 precursor pseudogene |
C00044491
|
| 725 | C4BPB, C4BP | complement component 4 binding protein, beta |
C00044491
|
| 55286 | C4orf19 | chromosome 4 open reading frame 19 |
C00044491
|
| 55345 | C4orf21 | chromosome 4 open reading frame 21 |
C00044491
|
| 132720 | C4orf32 | chromosome 4 open reading frame 32 |
C00044491
|
| 132321 | C4orf33 | chromosome 4 open reading frame 33 |
C00044491
|
| 56951 | C5orf15, KCT2 | chromosome 5 open reading frame 15 |
C00044491
|
| 439936 |
C00044491
|
||
| 64417 | C5orf28 | chromosome 5 open reading frame 28 |
C00044491
|
| 375444 | C5orf34 | chromosome 5 open reading frame 34 |
C00044491
|
| 65250 | C5orf42, JBTS17 | chromosome 5 open reading frame 42 |
C00044491
|
| 51149 | C5orf45 | chromosome 5 open reading frame 45 |
C00044491
|
| 644994 | C5orf50, SMIM23 | chromosome 5 open reading frame 50 |
C00044491
|
| 221491 | C6orf1, LBH | chromosome 6 open reading frame 1 |
C00044491
|
| 221545 | C6orf136 | chromosome 6 open reading frame 136 |
C00044491
|
| 29113 | C6orf15, STG | chromosome 6 open reading frame 15 |
C00044491
|
| 221416 | C6orf223 | chromosome 6 open reading frame 223 |
C00044491
|
| 57827 | C6orf47, D6S53E, G4, NG34 | chromosome 6 open reading frame 47 |
C00044491
|
| 50854 | C6orf48, D6S57, G8 | chromosome 6 open reading frame 48 |
C00044491
|
| 221477 | C6orf89, BRAP | chromosome 6 open reading frame 89 |
C00044491
|
| 730 | C7 | complement component 7 |
C00044491
|
| 78996 | C7orf49, MRI | chromosome 7 open reading frame 49 |
C00044491
|
| 203111 | C8orf47 | chromosome 8 open reading frame 47 |
C00044491
|
| 203076 | C8orf74 | chromosome 8 open reading frame 74 |
C00044491
|
| 79095 | C9orf16, EST00098 | chromosome 9 open reading frame 16 |
C00044491
|
| 389799 | C9orf171 | chromosome 9 open reading frame 171 |
C00044491
|
| 84909 | C9orf3, AOPEP, AP-O, APO, C90RF3, ONPEP | chromosome 9 open reading frame 3 |
C00044491
|
| 138199 | C9orf41 | chromosome 9 open reading frame 41 |
C00044491
|
| 203228 | C9orf72, ALSFTD, FTDALS | chromosome 9 open reading frame 72 |
C00044491
|
| 51759 | C9orf78, HCA59, HSPC220, bA409K20.3 | chromosome 9 open reading frame 78 |
C00044491
|
| 138241 | C9orf85 | chromosome 9 open reading frame 85 |
C00044491
|
| 766 | CA7, CAVII | carbonic anhydrase VII (EC:4.2.1.1) |
C00044491
|
| 81928 | CABLES2, C20orf150, dJ908M14.2, ik3-2 | Cdk5 and Abl enzyme substrate 2 |
C00044491
|
| 785 | CACNB4, CAB4, CACNLB4, EA5, EIG9, EJM, EJM4, EJM6 | calcium channel, voltage-dependent, beta 4 subunit |
C00044491
|
| 27101 | CACYBP, GIG5, RP1-102G20.6, S100A6BP, SIP | calcyclin binding protein |
C00044491
|
| 796 | CALCA, CALC1, CGRP, CGRP-I, CGRP1, CT, KC | calcitonin-related polypeptide alpha |
C00044491
|
| 797 | CALCB, CALC2, CGRP-II, CGRP2 | calcitonin-related polypeptide beta |
C00044491
|
| 10241 | CALCOCO2, NDP52 | calcium binding and coiled-coil domain 2 |
C00044491
|
| 800 | CALD1, CDM, H-CAD, HCAD, L-CAD, LCAD, NAG22 | caldesmon 1 |
C00044491
|
| 255022 | CALHM1, FAM26C | calcium homeostasis modulator 1 |
C00044491
|
| 91860 | CALML4, NY-BR-20 | calmodulin-like 4 |
C00044491
|
| 813 | CALU | calumenin |
C00044491
|
| 55450 | CAMK2N1, PRO1489, RP11-401M16.1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
C00044491
|
| 23261 | CAMTA1, CANPMR | calmodulin binding transcription activator 1 |
C00044491
|
| 23066 | CAND2, TIP120B, Tp120b | cullin-associated and neddylation-dissociated 2 (putative) |
C00044491
|
| 10487 | CAP1, CAP, CAP1-PEN | CAP, adenylate cyclase-associated protein 1 (yeast) |
C00044491
|
| 10486 | CAP2 | CAP, adenylate cyclase-associated protein, 2 (yeast) |
C00044491
|
| 823 | CAPN1, CANP, CANP1, CANPL1, muCANP, muCL | calpain 1, (mu/I) large subunit (EC:3.4.22.52) |
C00044491
|
| 6650 | CAPN15, SOLH | calpain 15 |
C00044491
|
| 824 | CAPN2, CANP2, CANPL2, CANPml, mCANP | calpain 2, (m/II) large subunit (EC:3.4.22.53) |
C00044491
|
| 4076 | CAPRIN1, GPIAP1, GPIP137, M11S1, RNG105, p137GPI | cell cycle associated protein 1 |
C00044491
|
| 65981 | CAPRIN2, C1QDC1, EEG-1, EEG1, RNG140 | caprin family member 2 |
C00044491
|
| 29775 | CARD10, BIMP1, CARMA3 | caspase recruitment domain family, member 10 |
C00044491
|
| 84674 | CARD6, CINCIN1 | caspase recruitment domain family, member 6 |
C00044491
|
| 10498 | CARM1, PRMT4 | coactivator-associated arginine methyltransferase 1 (EC:2.1.1.125) |
C00044491
|
| 57082 | CASC5, AF15Q14, CT29, D40, KNL1, PPP1R55, Spc7, hKNL-1, hSpc105 | cancer susceptibility candidate 5 |
C00044491
|
| 834 | CASP1, ICE, IL1BC, P45 | caspase 1, apoptosis-related cysteine peptidase (EC:3.4.22.36) |
C00044491
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00044491
|
| 837 | CASP4, ICE(rel)II, ICEREL-II, ICH-2, Mih1/TX, TX | caspase 4, apoptosis-related cysteine peptidase (EC:3.4.22.57) |
C00044491
|
| 839 | CASP6, MCH2 | caspase 6, apoptosis-related cysteine peptidase (EC:3.4.22.59) |
C00044491
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00044491
|
| 9994 | CASP8AP2, CED-4, FLASH, RIP25 | caspase 8 associated protein 2 |
C00044491
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00044491
|
| 857 | CAV1, BSCL3, CGL3, MSTP085, PPH3, VIP21, CAV | caveolin 1, caveolae protein, 22kDa |
C00044491
|
| 9139 | CBFA2T2, EHT, MTGR1, ZMYND3, p85 | core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
C00044491
|
| 865 | CBFB, PEBP2B | core-binding factor, beta subunit |
C00044491
|
| 867 | CBL, C-CBL, CBL2, FRA11B, NSLL, RNF55 | Cbl proto-oncogene, E3 ubiquitin protein ligase |
C00044491
|
| 873 | CBR1, CBR, SDR21C1, hCBR1 | carbonyl reductase 1 (EC:1.1.1.189 1.1.1.197 1.1.1.184) |
C00044491
|
| 874 | CBR3, SDR21C2, hCBR3 | carbonyl reductase 3 (EC:1.1.1.184) |
C00044491
|
| 84869 | CBR4, SDR45C1 | carbonyl reductase 4 (EC:1.1.1.-) |
C00044491
|
| 875 | CBS, HIP4 | cystathionine-beta-synthase (EC:4.2.1.22) |
C00044491
|
| 8535 | CBX4, NBP16, PC2 | chromobox homolog 4 |
C00044491
|
| 23468 | CBX5, HP1, HP1A | chromobox homolog 5 |
C00044491
|
| 23492 | CBX7 | chromobox homolog 7 |
C00044491
|
| 55610 | CCDC132 | coiled-coil domain containing 132 |
C00044491
|
| 64770 | CCDC14 | coiled-coil domain containing 14 |
C00044491
|
| 130940 | CCDC148 | coiled-coil domain containing 148 |
C00044491
|
| 284992 | CCDC150 | coiled-coil domain containing 150 |
C00044491
|
| 147872 | CCDC155, KASH5 | coiled-coil domain containing 155 (EC:3.1.3.48) |
C00044491
|
| 343099 | CCDC18, NY-SAR-41, dJ717I23.1 | coiled-coil domain containing 18 |
C00044491
|
| 25901 | CCDC28A, C6orf80, CCRL1AP | coiled-coil domain containing 28A |
C00044491
|
| 57003 | CCDC47, MSTP041 | coiled-coil domain containing 47 |
C00044491
|
| 152137 | CCDC50, C3orf6, DFNA44, YMER | coiled-coil domain containing 50 |
C00044491
|
| 79714 | CCDC51 | coiled-coil domain containing 51 |
C00044491
|
| 131076 | CCDC58 | coiled-coil domain containing 58 |
C00044491
|
| 29080 | CCDC59, BR22, TAP26 | coiled-coil domain containing 59 |
C00044491
|
| 84318 | CCDC77 | coiled-coil domain containing 77 |
C00044491
|
| 79080 | CCDC86 | coiled-coil domain containing 86 |
C00044491
|
| 54520 | CCDC93 | coiled-coil domain containing 93 |
C00044491
|
| 54535 | CCHCR1, C6orf18, HCR, SBP | coiled-coil alpha-helical rod protein 1 |
C00044491
|
| 885 | CCK | cholecystokinin |
C00044491
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00044491
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00044491
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00044491
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00044491
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00044491
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00044491
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00044491
|
| 57018 | CCNL1, ANIA6A, PRO1073, ania-6a | cyclin L1 |
C00044491
|
| 905 | CCNT2, CYCT2 | cyclin T2 |
C00044491
|
| 151195 | CCNYL1 | cyclin Y-like 1 |
C00044491
|
| 1230 | CCR1, CD191, CKR-1, CKR1, CMKBR1, HM145, MIP1aR, SCYAR1 | chemokine (C-C motif) receptor 1 |
C00044491
|
| 1235 | CCR6, BN-1, C-C_CKR-6, CC-CKR-6, CCR-6, CD196, CKR-L3, CKRL3, CMKBR6, DCR2, DRY6, GPR29, GPRCY4, STRL22 | chemokine (C-C motif) receptor 6 |
C00044491
|
| 25819 | CCRN4L, CCR4L, NOC | CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
C00044491
|
| 9973 | CCS | copper chaperone for superoxide dismutase |
C00044491
|
| 126731 | CCSAP, C1orf96, CSAP | centriole, cilia and spindle-associated protein |
C00044491
|
| 54462 | CCSER2, FAM190B, Gcap14, KIAA1128, bA486O22.1 | coiled-coil serine-rich protein 2 |
C00044491
|
| 908 | CCT6A, CCT-zeta, CCT-zeta-1, CCT6, Cctz, HTR3, MoDP-2, TCP-1-zeta, TCP20, TCPZ, TTCP20 | chaperonin containing TCP1, subunit 6A (zeta 1) |
C00044491
|
| 135228 | CD109, CPAMD7, RP11-525G3.1 | CD109 molecule |
C00044491
|
| 929 | CD14 | CD14 molecule |
C00044491
|
| 977 | CD151, GP27, MER2, PETA-3, RAPH, SFA1, TSPAN24 | CD151 molecule (Raph blood group) |
C00044491
|
| 909 | CD1A, CD1, FCB6, HTA1, R4, T6 | CD1a molecule |
C00044491
|
| 914 | CD2, LFA-2, SRBC, T11 | CD2 molecule |
C00044491
|
| 23607 | CD2AP, CMS | CD2-associated protein |
C00044491
|
| 146894 | CD300LG, CLM-9, CLM9, NEPMUCIN, TREM-4, TREM4 | CD300 molecule-like family member g |
C00044491
|
| 951 | CD37, GP52-40, TSPAN26 | CD37 molecule |
C00044491
|
| 915 | CD3D, CD3-DELTA, T3D | CD3d molecule, delta (CD3-TCR complex) |
C00044491
|
| 960 | CD44, CDW44, CSPG8, ECMR-III, HCELL, HUTCH-I, IN, LHR, MC56, MDU2, MDU3, MIC4, Pgp1 | CD44 molecule (Indian blood group) |
C00044491
|
| 962 | CD48, BCM1, BLAST, BLAST1, MEM-102, SLAMF2, hCD48, mCD48 | CD48 molecule |
C00044491
|
| 1604 | CD55, CR, CROM, DAF, TC | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
C00044491
|
| 965 | CD58, LFA-3, LFA3, ag3 | CD58 molecule |
C00044491
|
| 966 | CD59, 16.3A5, 1F5, EJ16, EJ30, EL32, G344, HRF-20, HRF20, MAC-IP, MACIF, MEM43, MIC11, MIN1, MIN2, MIN3, MIRL, MSK21, p18-20 | CD59 molecule, complement regulatory protein |
C00044491
|
| 969 | CD69, AIM, BL-AC/P26, CLEC2C, EA1, GP32/28, MLR-3 | CD69 molecule |
C00044491
|
| 973 | CD79A, IGA, MB-1 | CD79a molecule, immunoglobulin-associated alpha |
C00044491
|
| 975 | CD81, CVID6, S5.7, TAPA1, TSPAN28 | CD81 molecule |
C00044491
|
| 928 | CD9, BTCC-1, DRAP-27, MIC3, MRP-1, TSPAN-29, TSPAN29 | CD9 molecule |
C00044491
|
| 22918 | CD93, C1QR1, C1qR(P), C1qRP, CDw93, ECSM3, MXRA4, dJ737E23.1 | CD93 molecule |
C00044491
|
| 83692 | CD99L2, CD99B, MIC2L1 | CD99 molecule-like 2 |
C00044491
|
| 81602 | CDADC1, NYD-SP15, bA103J18.1 | cytidine and dCMP deaminase domain containing 1 |
C00044491
|
| 8872 | CDC123, C10orf7, D123 | cell division cycle 123 |
C00044491
|
| 34411 |
C00044491
|
||
| 8697 | CDC23, ANAPC8, APC8, CUT23 | cell division cycle 23 |
C00044491
|
| 993 | CDC25A, CDC25A2 | cell division cycle 25A (EC:3.1.3.48) |
C00044491
|
| 994 | CDC25B | cell division cycle 25B (EC:3.1.3.48) |
C00044491
|
| 996 | CDC27, ANAPC3, APC3, CDC27Hs, D0S1430E, D17S978E, HNUC, NUC2 | cell division cycle 27 |
C00044491
|
| 11140 | CDC37, P50CDC37 | cell division cycle 37 |
C00044491
|
| 55561 | CDC42BPG, DMPK2, HSMDPKIN, KAPPA-200, MRCKG, MRCKgamma | CDC42 binding protein kinase gamma (DMPK-like) (EC:2.7.11.1) |
C00044491
|
| 10602 | CDC42EP3, BORG2, CEP3, UB1 | CDC42 effector protein (Rho GTPase binding) 3 |
C00044491
|
| 56882 | CDC42SE1, SCIP1, SPEC1 | CDC42 small effector 1 |
C00044491
|
| 31052 |
C00044491
|
||
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00044491
|
| 157313 | CDCA2, Repo-Man | cell division cycle associated 2 |
C00044491
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00044491
|
| 55143 | CDCA8, BOR, BOREALIN, DasraB, MESRGP | cell division cycle associated 8 |
C00044491
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00044491
|
| 1002 | CDH4, CAD4, RCAD | cadherin 4, type 1, R-cadherin (retinal) |
C00044491
|
| 29965 | CDIP1, C16orf5, CDIP, I1, LITAFL | cell death-inducing p53 target 1 |
C00044491
|
| 8558 | CDK10, PISSLRE | cyclin-dependent kinase 10 (EC:2.7.11.22) |
C00044491
|
| 5218 | CDK14, PFTAIRE1, PFTK1 | cyclin-dependent kinase 14 (EC:2.7.11.22) |
C00044491
|
| 23097 | CDK19, CDC2L6, CDK11, bA346C16.3 | cyclin-dependent kinase 19 (EC:2.7.11.22) |
C00044491
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00044491
|
| 8941 | CDK5R2, NCK5AI, P39, p39nck5ai | cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
C00044491
|
| 55755 | CDK5RAP2, C48, Cep215, MCPH3 | CDK5 regulatory subunit associated protein 2 |
C00044491
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00044491
|
| 1025 | CDK9, C-2k, CDC2L4, CTK1, PITALRE, TAK | cyclin-dependent kinase 9 (EC:2.7.11.23 2.7.11.22) |
C00044491
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00044491
|
| 1028 | CDKN1C, BWCR, BWS, KIP2, WBS, p57, p57Kip2 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
C00044491
|
| 55602 | CDKN2AIP, CARF | CDKN2A interacting protein |
C00044491
|
| 1032 | CDKN2D, INK4D, p19, p19-INK4D | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
C00044491
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00044491
|
| 1036 | CDO1 | cysteine dioxygenase type 1 (EC:1.13.11.20) |
C00044491
|
| 1039 | CDR2, CDR62, Yo | cerebellar degeneration-related protein 2, 62kDa |
C00044491
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00044491
|
| 55573 | CDV3, H41 | CDV3 homolog (mouse) |
C00044491
|
| 634 | CEACAM1, BGP, BGP1, BGPI | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
C00044491
|
| 1089 | CEACAM4, CGM7, CGM7_HUMAN, NCA | carcinoembryonic antigen-related cell adhesion molecule 4 |
C00044491
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00044491
|
| 1054 | CEBPG, GPE1BP, IG/EBP-1 | CCAAT/enhancer binding protein (C/EBP), gamma |
C00044491
|
| 27440 | CECR5 | cat eye syndrome chromosome region, candidate 5 |
C00044491
|
| 1056 | CEL, BAL, BSDL, BSSL, CELL, CEase, FAP, FAPP, LIPA, MODY8 | carboxyl ester lipase (EC:3.1.1.13 3.1.1.3) |
C00044491
|
| 10658 | CELF1, BRUNOL2, CUG-BP, CUGBP, CUGBP1, EDEN-BP, NAB50, NAPOR, hNab50 | CUGBP, Elav-like family member 1 |
C00044491
|
| 56853 | CELF4, BRUNOL-4, BRUNOL4 | CUGBP, Elav-like family member 4 |
C00044491
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00044491
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00044491
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00044491
|
| 2491 | CENPI, CENP-I, FSHPRH1, LRPR1, Mis6 | centromere protein I |
C00044491
|
| 55835 | CENPJ, BM032, CENP-J, CPAP, LAP, LIP1, MCPH6, SASS4, SCKL4, Sas-4 | centromere protein J |
C00044491
|
| 64105 | CENPK, AF5alpha, CENP-K, P33, Solt | centromere protein K |
C00044491
|
| 55839 | CENPN, BM039, C16orf60, CENP-N, ICEN32 | centromere protein N |
C00044491
|
| 79172 | CENPO, CENP-O, ICEN-36, MCM21R | centromere protein O |
C00044491
|
| 201161 | CENPV, 3110013H01Rik, CENP-V, PRR6, p30 | centromere protein V |
C00044491
|
| 387103 | CENPW, C6orf173, CENP-W, CUG2 | centromere protein W |
C00044491
|
| 153241 | CEP120, CCDC100 | centrosomal protein 120kDa |
C00044491
|
| 145508 | CEP128, C14orf145, C14orf61, LEDP/132 | centrosomal protein 128kDa |
C00044491
|
| 22995 | CEP152, MCPH4, MCPH9, SCKL5 | centrosomal protein 152kDa |
C00044491
|
| 283638 | CEP170B, CEP170R, FAM68C, KIAA0284 | centrosomal protein 170B |
C00044491
|
| 55125 | CEP192, PPP1R62 | centrosomal protein 192kDa |
C00044491
|
| 11190 | CEP250, C-NAP1, CEP2, CNAP1 | centrosomal protein 250kDa |
C00044491
|
| 55165 | CEP55, C10orf3, CT111, URCC6 | centrosomal protein 55kDa |
C00044491
|
| 80321 | CEP70, BITE | centrosomal protein 70kDa |
C00044491
|
| 55722 | CEP72 | centrosomal protein 72kDa |
C00044491
|
| 79959 | CEP76, C18orf9, HsT1705 | centrosomal protein 76kDa |
C00044491
|
| 84131 | CEP78, C9orf81, IP63 | centrosomal protein 78kDa |
C00044491
|
| 64793 | CEP85, CCDC21 | centrosomal protein 85kDa |
C00044491
|
| 387119 | CEP85L, C6orf204, NY-BR-15, bA57K17.2 | centrosomal protein 85kDa-like |
C00044491
|
| 84902 | CEP89, CCDC123, CEP123 | centrosomal protein 89kDa |
C00044491
|
| 79598 | CEP97, 2810403B08Rik, LRRIQ2 | centrosomal protein 97kDa |
C00044491
|
| 64781 | CERK, LK4, dA59H18.2, dA59H18.3, hCERK | ceramide kinase (EC:2.7.1.138) |
C00044491
|
| 79603 | CERS4, LASS4, Trh1 | ceramide synthase 4 |
C00044491
|
| 253782 | CERS6, CERS5, LASS6 | ceramide synthase 6 |
C00044491
|
| 1066 | CES1, ACAT, CE-1, CEH, CES2, HMSE, HMSE1, PCE-1, REH, SES1, TGH, hCE-1 | carboxylesterase 1 (EC:3.1.1.1 3.1.1.56) |
C00044491
|
| 8824 | CES2, CE-2, CES2A1, PCE-2, iCE | carboxylesterase 2 (EC:3.1.1.1 3.1.1.84 3.1.1.56) |
C00044491
|
| 1073 | CFL2, NEM7 | cofilin 2 (muscle) |
C00044491
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00044491
|
| 8545 | CGGBP1, CGGBP, p20-CGGBP | CGG triplet repeat binding protein 1 |
C00044491
|
| 57530 | CGN | cingulin |
C00044491
|
| 10669 | CGREF1, CGR11 | cell growth regulator with EF-hand domain 1 |
C00044491
|
| 79094 | CHAC1 | ChaC, cation transport regulator homolog 1 (E. coli) |
C00044491
|
| 1101 | CHAD, SLRR4A | chondroadherin |
C00044491
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00044491
|
| 8208 | CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7, MPP7 | chromatin assembly factor 1, subunit B (p60) |
C00044491
|
| 79145 | CHCHD7, COX23 | coiled-coil-helix-coiled-coil-helix domain containing 7 |
C00044491
|
| 1108 | CHD4, Mi-2b, Mi2-BETA | chromodomain helicase DNA binding protein 4 (EC:3.6.4.12) |
C00044491
|
| 55636 | CHD7, HH5, IS3, KAL5 | chromodomain helicase DNA binding protein 7 (EC:3.6.4.12) |
C00044491
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00044491
|
| 5119 | CHMP1A, CHMP1, PCH8, PCOLN3, PRSM1, VPS46-1, VPS46A | charged multivesicular body protein 1A |
C00044491
|
| 1124 | CHN2, ARHGAP3, BCH, CHN2-3, RHOGAP3 | chimerin 2 |
C00044491
|
| 26973 | CHORDC1, CHP1 | cysteine and histidine-rich domain (CHORD) containing 1 |
C00044491
|
| 8973 | CHRNA6, CHNRA6 | cholinergic receptor, nicotinic, alpha 6 (neuronal) |
C00044491
|
| 1144 | CHRND, ACHRD, CMS2A, FCCMS, SCCMS | cholinergic receptor, nicotinic, delta (muscle) |
C00044491
|
| 9435 | CHST2, C6ST, GST-2, GST2, Gn6ST-1, glcNAc6ST-1 | carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
C00044491
|
| 4166 | CHST6, MCDC1 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
C00044491
|
| 64377 | CHST8, GALNAC4ST1, GalNAc4ST | carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8 |
C00044491
|
| 63922 | CHTF18, C16orf41, C321D2.2, C321D2.3, C321D2.4, CHL12, Ctf18, RUVBL | CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
C00044491
|
| 1153 | CIRBP, CIRP | cold inducible RNA binding protein |
C00044491
|
| 84916 | CIRH1A, CIRHIN, NAIC, TEX292, UTP4 | cirrhosis, autosomal recessive 1A (cirhin) |
C00044491
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00044491
|
| 26586 | CKAP2, LB1, TMAP, se20-10 | cytoskeleton associated protein 2 |
C00044491
|
| 150468 | CKAP2L | cytoskeleton associated protein 2-like |
C00044491
|
| 10970 | CKAP4, CLIMP-63, ERGIC-63, p63 | cytoskeleton-associated protein 4 |
C00044491
|
| 9793 | CKAP5, CHTOG, MSPS, TOG, TOGp, ch-TOG | cytoskeleton associated protein 5 |
C00044491
|
| 51192 | CKLF, C32, CKLF1, CKLF2, CKLF3, CKLF4, UCK-1 | chemokine-like factor |
C00044491
|
| 1158 | CKM, CKMM, M-CK | creatine kinase, muscle (EC:2.7.3.2) |
C00044491
|
| 1163 | CKS1B, CKS1, PNAS-16, PNAS-18, ckshs1 | CDC28 protein kinase regulatory subunit 1B |
C00044491
|
| 1164 | CKS2, CKSHS2 | CDC28 protein kinase regulatory subunit 2 |
C00044491
|
| 23155 | CLCC1, MCLC | chloride channel CLIC-like 1 |
C00044491
|
| 1182 | CLCN3, CLC3, ClC-3 | chloride channel, voltage-sensitive 3 |
C00044491
|
| 9076 | CLDN1, CLD1, ILVASC, SEMP1 | claudin 1 |
C00044491
|
| 9069 | CLDN12 | claudin 12 |
C00044491
|
| 9075 | CLDN2 | claudin 2 |
C00044491
|
| 137075 | CLDN23, CLDNL, hCG1646163 | claudin 23 |
C00044491
|
| 9080 | CLDN9 | claudin 9 |
C00044491
|
| 56650 | CLDND1, C3orf4, GENX-3745 | claudin domain containing 1 |
C00044491
|
| 10462 | CLEC10A, CD301, CLECSF13, CLECSF14, HML, HML2, MGL | C-type lectin domain family 10, member A |
C00044491
|
| 23274 | CLEC16A, Gop-1, KIAA0350 | C-type lectin domain family 16, member A |
C00044491
|
| 64581 | CLEC7A, BGR, CANDF4, CLECSF12, DECTIN1 | C-type lectin domain family 7, member A |
C00044491
|
| 1192 | CLIC1, G6, NCC27 | chloride intracellular channel 1 |
C00044491
|
| 9022 | CLIC3 | chloride intracellular channel 3 |
C00044491
|
| 53405 | CLIC5, MST130, MSTP130 | chloride intracellular channel 5 |
C00044491
|
| 9685 | CLINT1, CLINT, ENTH, EPN4, EPNR | clathrin interactor 1 |
C00044491
|
| 6249 | CLIP1, CLIP, CLIP-170, CLIP170, CYLN1, RSN | CAP-GLY domain containing linker protein 1 |
C00044491
|
| 25999 | CLIP3, CLIPR-59, CLIPR59, RSNL1 | CAP-GLY domain containing linker protein 3 |
C00044491
|
| 1195 | CLK1, CLK, CLK/STY, STY | CDC-like kinase 1 (EC:2.7.12.1) |
C00044491
|
| 79789 | CLMN | calmin (calponin-like, transmembrane) |
C00044491
|
| 2055 | CLN8, C8orf61, EPMR | ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) |
C00044491
|
| 1207 | CLNS1A, CLCI, CLNS1B, ICln | chloride channel, nucleotide-sensitive, 1A |
C00044491
|
| 1209 | CLPTM1 | cleft lip and palate associated transmembrane protein 1 |
C00044491
|
| 10845 | CLPX | caseinolytic mitochondrial matrix peptidase chaperone subunit |
C00044491
|
| 63967 | CLSPN | claspin |
C00044491
|
| 22883 | CLSTN1, ALC-ALPHA, CDHR12, CSTN1, PIK3CD, XB31alpha, alcalpha1, alcalpha2 | calsyntenin 1 |
C00044491
|
| 1213 | CLTC, CHC, CHC17, CLH-17, CLTCL2, Hc | clathrin, heavy chain (Hc) |
C00044491
|
| 23277 | CLUH, CLU1, KIAA0664 | clustered mitochondria (cluA/CLU1) homolog |
C00044491
|
| 134147 | CMBL, JS-1 | carboxymethylenebutenolidase homolog (Pseudomonas) (EC:3.1.-.-) |
C00044491
|
| 129607 | CMPK2, TMPK2, TYKi, UMP-CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial (EC:2.7.4.6 2.7.4.14) |
C00044491
|
| 146225 | CMTM2, CKLFSF2 | CKLF-like MARVEL transmembrane domain containing 2 |
C00044491
|
| 146223 | CMTM4, CKLFSF4 | CKLF-like MARVEL transmembrane domain containing 4 |
C00044491
|
| 23070 | CMTR1, FTSJD2, KIAA0082, MTr1, hMTr1 | cap methyltransferase 1 (EC:2.1.1.57) |
C00044491
|
| 55783 | CMTR2, AFT, FTSJD1, HMTr2, MTr2 | cap methyltransferase 2 |
C00044491
|
| 84735 | CNDP1, CN1, CPGL2, HsT2308 | carnosine dipeptidase 1 (metallopeptidase M20 family) (EC:3.4.13.20) |
C00044491
|
| 55748 | CNDP2, CN2, CPGL, HsT2298, PEPA | CNDP dipeptidase 2 (metallopeptidase M20 family) (EC:3.4.13.18) |
C00044491
|
| 255919 | CNEP1R1, C16orf69, NEP1-R1, TMEM188, TMP125 | CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
C00044491
|
| 149111 | CNIH3, CNIH-3 | cornichon family AMPA receptor auxiliary protein 3 |
C00044491
|
| 26504 | CNNM4, ACDP4 | cyclin M4 |
C00044491
|
| 55571 | CNOT11, C2orf29 | CCR4-NOT transcription complex, subunit 11 |
C00044491
|
| 29883 | CNOT7, CAF1, Caf1a, hCAF-1 | CCR4-NOT transcription complex, subunit 7 (EC:3.1.13.4) |
C00044491
|
| 10695 | CNPY3, CAG4A, ERDA5, PRAT4A, TNRC5 | canopy FGF signaling regulator 3 |
C00044491
|
| 25927 | CNRIP1, C2orf32, CRIP1 | cannabinoid receptor interacting protein 1 |
C00044491
|
| 5067 | CNTN3, BIG-1, PANG, PCS | contactin 3 (plasmacytoma associated) |
C00044491
|
| 116840 | CNTROB, LIP8 | centrobin, centrosomal BRCA2 interacting protein |
C00044491
|
| 80347 | COASY, DPCK, NBP, PPAT, UKR1, pOV-2 | CoA synthase (EC:2.7.7.3 2.7.1.24) |
C00044491
|
| 23242 | COBL | cordon-bleu WH2 repeat protein |
C00044491
|
| 1690 | COCH, COCH-5B2, COCH5B2, DFNA9, DFNA31 | cochlin |
C00044491
|
| 83548 | COG3, SEC34 | component of oligomeric golgi complex 3 |
C00044491
|
| 25839 | COG4, CDG2J, COD1 | component of oligomeric golgi complex 4 |
C00044491
|
| 10466 | COG5, CDG2I, GOLTC1, GTC90 | component of oligomeric golgi complex 5 |
C00044491
|
| 57511 | COG6, CDG2L, COD2, SHNS | component of oligomeric golgi complex 6 |
C00044491
|
| 91949 | COG7, CDG2E | component of oligomeric golgi complex 7 |
C00044491
|
| 8161 | COIL, CLN80, p80-coilin | coilin |
C00044491
|
| 1305 | COL13A1, COLXIIIA1 | collagen, type XIII, alpha 1 |
C00044491
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00044491
|
| 1278 | COL1A2, OI4 | collagen, type I, alpha 2 |
C00044491
|
| 85301 | COL27A1 | collagen, type XXVII, alpha 1 |
C00044491
|
| 1281 | COL3A1, EDS4A | collagen, type III, alpha 1 |
C00044491
|
| 1286 | COL4A4, CA44 | collagen, type IV, alpha 4 |
C00044491
|
| 1290 | COL5A2 | collagen, type V, alpha 2 |
C00044491
|
| 1293 | COL6A3 | collagen, type VI, alpha 3 |
C00044491
|
| 1294 | COL7A1, EBD1, EBDCT, EBR1 | collagen, type VII, alpha 1 |
C00044491
|
| 81035 | COLEC12, CLP1, NSR2, SCARA4, SRCL | collectin sub-family member 12 |
C00044491
|
| 51122 | COMMD2 | COMM domain containing 2 |
C00044491
|
| 23412 | COMMD3, BUP, C10orf8 | COMM domain containing 3 |
C00044491
|
| 1311 | COMP, EDM1, EPD1, MED, PSACH, THBS5 | cartilage oligomeric matrix protein |
C00044491
|
| 118881 | COMTD1 | catechol-O-methyltransferase domain containing 1 |
C00044491
|
| 1314 | COPA, HEP-COP | coatomer protein complex, subunit alpha |
C00044491
|
| 10987 | COPS5, CSN5, JAB1, MOV-34, SGN5 | COP9 signalosome subunit 5 |
C00044491
|
| 10920 | COPS8, COP9, CSN8, SGN8 | COP9 signalosome subunit 8 |
C00044491
|
| 80219 | COQ10B | coenzyme Q10 homolog B (S. cerevisiae) |
C00044491
|
| 57175 | CORO1B, CORONIN-2 | coronin, actin binding protein, 1B |
C00044491
|
| 1355 | COX15, CEMCOX2 | cytochrome c oxidase assembly homolog 15 (yeast) |
C00044491
|
| 285521 | COX18, COX18HS | COX18 cytochrome C oxidase assembly factor |
C00044491
|
| 1349 | COX7B, APLCC | cytochrome c oxidase subunit VIIb (EC:1.9.3.1) |
C00044491
|
| 1351 | COX8A, COX, COX8, COX8-2, COX8L, VIII, VIII-L | cytochrome c oxidase subunit VIIIA (ubiquitous) (EC:1.9.3.1) |
C00044491
|
| 1356 | CP, CP-2 | ceruloplasmin (ferroxidase) (EC:1.16.3.1) |
C00044491
|
| 33136 |
C00044491
|
||
| 1361 | CPB2, CPU, PCPB, TAFI | carboxypeptidase B2 (plasma) (EC:3.4.17.20) |
C00044491
|
| 80315 | CPEB4 | cytoplasmic polyadenylation element binding protein 4 |
C00044491
|
| 79974 | CPED1, C7orf58 | cadherin-like and PC-esterase domain containing 1 |
C00044491
|
| 10815 | CPLX1, CPX-I, CPX1 | complexin 1 |
C00044491
|
| 1368 | CPM | carboxypeptidase M (EC:3.4.17.12) |
C00044491
|
| 1370 | CPN2, ACBP | carboxypeptidase N, polypeptide 2 (EC:3.4.17.3) |
C00044491
|
| 221184 | CPNE2, COPN2, CPN2 | copine II |
C00044491
|
| 53981 | CPSF2, CPSF100 | cleavage and polyadenylation specific factor 2, 100kDa |
C00044491
|
| 51692 | CPSF3, CPSF-73, CPSF73 | cleavage and polyadenylation specific factor 3, 73kDa |
C00044491
|
| 11052 | CPSF6, CFIM, CFIM68, HPBRII-4, HPBRII-7 | cleavage and polyadenylation specific factor 6, 68kDa |
C00044491
|
| 1376 | CPT2, CPT1, CPTASE, IIAE4 | carnitine palmitoyltransferase 2 (EC:2.3.1.21) |
C00044491
|
| 54504 | CPVL, HVLP | carboxypeptidase, vitellogenic-like |
C00044491
|
| 8532 | CPZ | carboxypeptidase Z |
C00044491
|
| 84699 | CREB3L3, CREB-H, CREBH | cAMP responsive element binding protein 3-like 3 |
C00044491
|
| 1390 | CREM, CREM-2, ICER, hCREM-2 | cAMP responsive element modulator |
C00044491
|
| 51232 | CRIM1, CRIM-1, S52 | cysteine rich transmembrane BMP regulator 1 (chordin-like) |
C00044491
|
| 100189550 |
C00044491
|
||
| 1397 | CRIP2, CRIP, CRP2, ESP1 | cysteine-rich protein 2 |
C00044491
|
| 285464 | CRIPAK | cysteine-rich PAK1 inhibitor |
C00044491
|
| 1398 | CRK, CRKII, p38 | v-crk avian sarcoma virus CT10 oncogene homolog |
C00044491
|
| 49860 | CRNN, C1orf10, DRC1, PDRC1, SEP53 | cornulin |
C00044491
|
| 84809 | CROCCP2, CROCCL1 | ciliary rootlet coiled-coil, rootletin pseudogene 2 |
C00044491
|
| 64784 | CRTC3, TORC-3, TORC3 | CREB regulated transcription coactivator 3 |
C00044491
|
| 1407 | CRY1, PHLL1 | cryptochrome 1 (photolyase-like) |
C00044491
|
| 1408 | CRY2, HCRY2, PHLL2 | cryptochrome 2 (photolyase-like) |
C00044491
|
| 1410 | CRYAB, CMD1II, CRYA2, CTPP2, CTRCT16, HSPB5, MFM2 | crystallin, alpha B |
C00044491
|
| 1413 | CRYBA4, CTRCT23, MCOPCT4 | crystallin, beta A4 |
C00044491
|
| 1429 | CRYZ | crystallin, zeta (quinone reductase) (EC:1.6.5.5) |
C00044491
|
| 1440 | CSF3, C17orf33, CSF3OS, GCSF, G-CSF | colony stimulating factor 3 (granulocyte) |
C00044491
|
| 1441 | CSF3R, CD114, GCSFR | colony stimulating factor 3 receptor (granulocyte) |
C00044491
|
| 55454 | CSGALNACT2, CHGN2, GALNACT-2, GALNACT2 | chondroitin sulfate N-acetylgalactosaminyltransferase 2 (EC:2.4.1.174) |
C00044491
|
| 1453 | CSNK1D, ASPS, CKIdelta, FASPS2, HCKID | casein kinase 1, delta (EC:2.7.11.1 2.7.11.26) |
C00044491
|
| 53944 | CSNK1G1, CK1gamma1 | casein kinase 1, gamma 1 (EC:2.7.11.1) |
C00044491
|
| 64651 | CSRNP1, AXUD1, CSRNP-1, FAM130B, TAIP-3, URAX1 | cysteine-serine-rich nuclear protein 1 |
C00044491
|
| 81566 | CSRNP2, C12orf2, C12orf22, FAM130A1, PPP1R72, TAIP-12 | cysteine-serine-rich nuclear protein 2 |
C00044491
|
| 1466 | CSRP2, CRP2, LMO5, SmLIM | cysteine and glycine-rich protein 2 |
C00044491
|
| 57325 | CSRP2BP, ATAC2, CRP2BP, KAT14, PRO1194, dJ717M23.1 | CSRP2 binding protein |
C00044491
|
| 1475 | CSTA, AREI, STF1, STFA | cystatin A (stefin A) |
C00044491
|
| 1478 | CSTF2, CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
C00044491
|
| 23283 | CSTF2T, CstF-64T | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
C00044491
|
| 1487 | CTBP1, BARS | C-terminal binding protein 1 |
C00044491
|
| 80169 | CTC1, AAF-132, AAF132, C17orf68, CRMCC, tmp494178 | CTS telomere maintenance complex component 1 |
C00044491
|
| 9150 | CTDP1, CCFDN, FCP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 (EC:3.1.3.16) |
C00044491
|
| 58190 | CTDSP1, NIF3, NLI-IF, NLIIF, SCP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 (EC:3.1.3.16) |
C00044491
|
| 10106 | CTDSP2, OS4, PSR2, SCP2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (EC:3.1.3.16) |
C00044491
|
| 10217 | CTDSPL, C3orf8, HYA22, PSR1, RBSP3, SCP3 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (EC:3.1.3.16) |
C00044491
|
| 51496 | CTDSPL2, HSPC129 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 (EC:3.1.3.-) |
C00044491
|
| 1490 | CTGF, CCN2, HCS24, IGFBP8, NOV2 | connective tissue growth factor |
C00044491
|
| 1491 | CTH | cystathionase (cystathionine gamma-lyase) (EC:4.4.1.1) |
C00044491
|
| 1496 | CTNNA2, CAP-R, CAPR, CT114, CTNR | catenin (cadherin-associated protein), alpha 2 |
C00044491
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00044491
|
| 1500 | CTNND1, CAS, CTNND, P120CAS, P120CTN, p120, p120(CAS), p120(CTN) | catenin (cadherin-associated protein), delta 1 |
C00044491
|
| 1501 | CTNND2, GT24, NPRAP | catenin (cadherin-associated protein), delta 2 |
C00044491
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00044491
|
| 1075 | CTSC, CPPI, DPP-I, DPP1, DPPI, HMS, JP, JPD, PALS, PDON1, PLS | cathepsin C (EC:3.4.14.1) |
C00044491
|
| 1512 | CTSH, ACC-4, ACC-5, CPSB, minichain | cathepsin H (EC:3.4.22.16) |
C00044491
|
| 1520 | CTSS | cathepsin S (EC:3.4.22.27) |
C00044491
|
| 1515 | CTSV, CATL2, CTSL2, CTSU | cathepsin V (EC:3.4.22.43) |
C00044491
|
| 2017 | CTTN, EMS1 | cortactin |
C00044491
|
| 90353 | CTU1, ATPBD3, NCS6 | cytosolic thiouridylase subunit 1 |
C00044491
|
| 8454 | CUL1 | cullin 1 |
C00044491
|
| 8451 | CUL4A | cullin 4A |
C00044491
|
| 8450 | CUL4B, MRXHF2, MRXS15, MRXSC, SFM2 | cullin 4B |
C00044491
|
| 8065 | CUL5, VACM-1, VACM1 | cullin 5 |
C00044491
|
| 1523 | CUX1, CASP, CDP, CDP/Cut, CDP1, COY1, CUTL1, CUX, Clox, Cux/CDP, GOLIM6, Nbla10317, p100, p110, p200, p75 | cut-like homeobox 1 |
C00044491
|
| 54883 | CWC25, CCDC49 | CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
C00044491
|
| 10283 | CWC27, NY-CO-10, SDCCAG10 | CWC27 spliceosome-associated protein homolog (S. cerevisiae) (EC:5.2.1.8) |
C00044491
|
| 653108 |
C00044491
|
||
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00044491
|
| 2920 | CXCL2, CINC-2a, GRO2, GROb, MGSA-b, MIP-2a, MIP2, MIP2A, SCYB2 | chemokine (C-X-C motif) ligand 2 |
C00044491
|
| 170063 | CXorf22 | chromosome X open reading frame 22 |
C00044491
|
| 203414 |
C00044491
|
||
| 51523 | CXXC5, CF5, RINF, WID | CXXC finger protein 5 |
C00044491
|
| 220002 | CYB561A3, CYBASC3, LCYTB | cytochrome b561 family, member A3 |
C00044491
|
| 11068 | CYB561D2, 101F6, TSP10 | cytochrome b561 family, member D2 |
C00044491
|
| 80777 | CYB5B, CYB5-M, CYPB5M, OMB5 | cytochrome b5 type B (outer mitochondrial membrane) |
C00044491
|
| 124637 | CYB5D1 | cytochrome b5 domain containing 1 |
C00044491
|
| 124936 | CYB5D2 | cytochrome b5 domain containing 2 |
C00044491
|
| 51706 | CYB5R1, B5R.1, B5R1, B5R2, NQO3A2, humb5R2 | cytochrome b5 reductase 1 (EC:1.6.2.2) |
C00044491
|
| 1537 | CYC1, MC3DN6, UQCR4 | cytochrome c-1 |
C00044491
|
| 360159 |
C00044491
|
||
| 26999 | CYFIP2, PIR121 | cytoplasmic FMR1 interacting protein 2 |
C00044491
|
| 114757 | CYGB, HGB, STAP | cytoglobin |
C00044491
|
| 50626 | CYHR1, CHRP | cysteine/histidine-rich 1 |
C00044491
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00044491
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00044491
|
| 57404 | CYP20A1, CYP-M | cytochrome P450, family 20, subfamily A, polypeptide 1 |
C00044491
|
| 1589 | CYP21A2, CA21H, CAH1, CPS1, CYP21, CYP21B, P450c21B | cytochrome P450, family 21, subfamily A, polypeptide 2 (EC:1.14.99.10) |
C00044491
|
| 1592 | CYP26A1, CP26, CYP26, P450RAI, P450RAI1 | cytochrome P450, family 26, subfamily A, polypeptide 1 (EC:1.14.-.-) |
C00044491
|
| 1593 | CYP27A1, CP27, CTX, CYP27 | cytochrome P450, family 27, subfamily A, polypeptide 1 (EC:1.14.13.15) |
C00044491
|
| 1548 | CYP2A6, CPA6, CYP2A, CYP2A3, CYPIIA6, P450C2A, P450PB | cytochrome P450, family 2, subfamily A, polypeptide 6 (EC:1.14.14.1) |
C00044491
|
| 1562 | CYP2C18, CPCI, CYP2C, CYP2C17, P450-6B/29C, P450IIC17 | cytochrome P450, family 2, subfamily C, polypeptide 18 (EC:1.14.14.1) |
C00044491
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00044491
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00044491
|
| 554227 |
C00044491
|
||
| 113612 | CYP2U1, P450TEC, SPG49, SPG56 | cytochrome P450, family 2, subfamily U, polypeptide 1 (EC:1.14.14.1) |
C00044491
|
| 54905 | CYP2W1 | cytochrome P450, family 2, subfamily W, polypeptide 1 |
C00044491
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00044491
|
| 64816 | CYP3A43 | cytochrome P450, family 3, subfamily A, polypeptide 43 (EC:1.14.14.1) |
C00044491
|
| 1551 | CYP3A7, CP37, CYPIIIA7, P450-HFLA | cytochrome P450, family 3, subfamily A, polypeptide 7 (EC:1.14.14.1) |
C00044491
|
| 66002 | CYP4F12, F22329_1 | cytochrome P450, family 4, subfamily F, polypeptide 12 (EC:1.14.14.1) |
C00044491
|
| 4051 | CYP4F3, CPF3, CYP4F, LTB4H | cytochrome P450, family 4, subfamily F, polypeptide 3 (EC:1.14.13.30) |
C00044491
|
| 1595 | CYP51A1, CP51, CYP51, CYPL1, LDM, P450-14DM, P450L1 | cytochrome P450, family 51, subfamily A, polypeptide 1 (EC:1.14.13.70) |
C00044491
|
| 3491 | CYR61, CCN1, GIG1, IGFBP10 | cysteine-rich, angiogenic inducer, 61 |
C00044491
|
| 9267 | CYTH1, B2-1, CYTOHESIN-1, D17S811E, PSCD1, SEC7 | cytohesin 1 |
C00044491
|
| 9266 | CYTH2, ARNO, CTS18, CTS18.1, PSCD2, PSCD2L, SEC7L, Sec7p-L, Sec7p-like | cytohesin 2 |
C00044491
|
| 9265 | CYTH3, ARNO3, GRP1, PSCD3 | cytohesin 3 |
C00044491
|
| 23002 | DAAM1 | dishevelled associated activator of morphogenesis 1 |
C00044491
|
| 1603 | DAD1, OST2 | defender against cell death 1 (EC:2.4.99.18) |
C00044491
|
| 1605 | DAG1, 156DAG, A3a, AGRNR, DAG, MDDGC7, MDDGC9 | dystroglycan 1 (dystrophin-associated glycoprotein 1) |
C00044491
|
| 26007 | DAK, NET45 | dihydroxyacetone kinase 2 homolog (S. cerevisiae) (EC:2.7.1.29 4.6.1.15 2.7.1.28) |
C00044491
|
| 1610 | DAO, DAAO, DAMOX, OXDA | D-amino-acid oxidase (EC:1.4.3.3) |
C00044491
|
| 1612 | DAPK1, DAPK | death-associated protein kinase 1 (EC:2.7.11.1) |
C00044491
|
| 1615 | DARS, HBSL | aspartyl-tRNA synthetase (EC:6.1.1.12) |
C00044491
|
| 1616 | DAXX, BING2, DAP6, EAP1 | death-domain associated protein |
C00044491
|
| 1618 | DAZL, DAZH, DAZL1, DAZLA, SPGYLA | deleted in azoospermia-like |
C00044491
|
| 80174 | DBF4B, ASKL1, CHIFB, DRF1, ZDBF1B | DBF4 homolog B (S. cerevisiae) |
C00044491
|
| 1622 | DBI, ACBD1, ACBP, CCK-RP, EP | diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) (EC:3.4.17.3) |
C00044491
|
| 1629 | DBT, BCATE2, BCKAD-E2, BCKADE2, E2, E2B | dihydrolipoamide branched chain transacylase E2 (EC:2.3.1.168) |
C00044491
|
| 25879 | DCAF13, GM83, WDSOF1 | DDB1 and CUL4 associated factor 13 |
C00044491
|
| 285429 | DCAF4L1, WDR21B | DDB1 and CUL4 associated factor 4-like 1 |
C00044491
|
| 55827 | DCAF6, 1200006M05Rik, ARCAP, IQWD1, MSTP055, NRIP, PC326 | DDB1 and CUL4 associated factor 6 |
C00044491
|
| 50717 | DCAF8, H326, WDR42A | DDB1 and CUL4 associated factor 8 |
C00044491
|
| 79877 | DCAKD | dephospho-CoA kinase domain containing |
C00044491
|
| 131566 | DCBLD2, CLCP1, ESDN | discoidin, CUB and LCCL domain containing 2 |
C00044491
|
| 64858 | DCLRE1B, APOLLO, SNM1B, SNMIB | DNA cross-link repair 1B |
C00044491
|
| 64421 | DCLRE1C, A-SCID, DCLREC1C, RS-SCID, SCIDA, SNM1C, hSNM1C | DNA cross-link repair 1C |
C00044491
|
| 196513 | DCP1B, DCP1, hDcp1b | decapping mRNA 1B |
C00044491
|
| 28960 | DCPS, DCS1, HINT-5, HINT5, HSL1 | decapping enzyme, scavenger (EC:3.6.1.59) |
C00044491
|
| 79077 | DCTPP1, RS21C6, XTP3TPA | dCTP pyrophosphatase 1 (EC:3.6.1.12) |
C00044491
|
| 23142 | DCUN1D4 | DCN1, defective in cullin neddylation 1, domain containing 4 |
C00044491
|
| 51181 | DCXR, DCR, HCR2, HCRII, KIDCR, P34H, SDR20C1, XR | dicarbonyl/L-xylulose reductase (EC:1.1.1.10) |
C00044491
|
| 1642 | DDB1, DDBA, UV-DDB1, XAP1, XPCE, XPE, XPE-BF | damage-specific DNA binding protein 1, 127kDa |
C00044491
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00044491
|
| 80821 | DDHD1, PA-PLA1, PAPLA1, SPG28 | DDHD domain containing 1 |
C00044491
|
| 23259 | DDHD2, SAMWD1, SPG54 | DDHD domain containing 2 |
C00044491
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00044491
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00044491
|
| 23109 | DDN | dendrin |
C00044491
|
| 10521 | DDX17, P72, RH70 | DEAD (Asp-Glu-Ala-Asp) box helicase 17 (EC:3.6.4.13) |
C00044491
|
| 8886 | DDX18, MrDb | DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 (EC:3.6.4.13) |
C00044491
|
| 11218 | DDX20, DP103, GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 (EC:3.6.4.13) |
C00044491
|
| 9416 | DDX23, PRPF28, SNRNP100, U5-100K, U5-100KD, prp28 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 (EC:3.6.4.13) |
C00044491
|
| 55794 | DDX28, MDDX28 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (EC:3.6.4.13) |
C00044491
|
| 54606 | DDX56, DDX21, DDX26, NOH61 | DEAD (Asp-Glu-Ala-Asp) box helicase 56 (EC:3.6.4.13) |
C00044491
|
| 23586 | DDX58, RIG-I, RIGI, RLR-1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (EC:3.6.4.13) |
C00044491
|
| 55601 | DDX60 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (EC:3.6.4.13) |
C00044491
|
| 91351 | DDX60L | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like (EC:3.6.4.13) |
C00044491
|
| 162989 | DEDD2, FLAME-3 | death effector domain containing 2 |
C00044491
|
| 245929 | DEFB115, DEFB-15 | defensin, beta 115 |
C00044491
|
| 8560 | DEGS1, DEGS, DEGS-1, DES1, Des-1, FADS7, MLD | delta(4)-desaturase, sphingolipid 1 (EC:1.14.-.-) |
C00044491
|
| 123099 | DEGS2, C14orf66, DES2, FADS8 | delta(4)-desaturase, sphingolipid 2 |
C00044491
|
| 163486 | DENND1B, C1ORF18, C1orf218, FAM31B | DENN/MADD domain containing 1B |
C00044491
|
| 163259 | DENND2C, dJ1156J9.1 | DENN/MADD domain containing 2C |
C00044491
|
| 79961 | DENND2D | DENN/MADD domain containing 2D |
C00044491
|
| 10260 | DENND4A, IRLB, MYCPBP | DENN/MADD domain containing 4A |
C00044491
|
| 9909 | DENND4B, KIAA0476 | DENN/MADD domain containing 4B |
C00044491
|
| 160518 | DENND5B | DENN/MADD domain containing 5B |
C00044491
|
| 8562 | DENR, DRP, DRP1, SMAP-3 | density-regulated protein |
C00044491
|
| 9681 | DEPDC5, DEP.5, FFEVF | DEP domain containing 5 |
C00044491
|
| 51029 | DESI2, C1orf121, DESI, DESI1, DeSI-2, FAM152A, PNAS-4, PPPDE1 | desumoylating isopeptidase 2 |
C00044491
|
| 1676 | DFFA, DFF-45, DFF1, ICAD | DNA fragmentation factor, 45kDa, alpha polypeptide |
C00044491
|
| 1687 | DFNA5, ICERE-1 | deafness, autosomal dominant 5 |
C00044491
|
| 8694 | DGAT1, ARAT, ARGP1, DGAT | diacylglycerol O-acyltransferase 1 (EC:2.3.1.20 2.3.1.76) |
C00044491
|
| 84649 | DGAT2, ARAT, GS1999FULL | diacylglycerol O-acyltransferase 2 (EC:2.3.1.20 2.3.1.76) |
C00044491
|
| 1606 | DGKA, DAGK, DAGK1, DGK-alpha | diacylglycerol kinase, alpha 80kDa (EC:2.7.1.107) |
C00044491
|
| 8527 | DGKD, DGKdelta, dgkd-2 | diacylglycerol kinase, delta 130kDa (EC:2.7.1.107) |
C00044491
|
| 8525 | DGKZ, DAGK5, DAGK6, DGK-ZETA, hDGKzeta | diacylglycerol kinase, zeta (EC:2.7.1.107) |
C00044491
|
| 1716 | DGUOK, MTDPS3, dGK | deoxyguanosine kinase (EC:2.7.1.113) |
C00044491
|
| 1717 | DHCR7, SLOS | 7-dehydrocholesterol reductase (EC:1.3.1.21) |
C00044491
|
| 1719 | DHFR, DHFRP1, DYR | dihydrofolate reductase (EC:1.5.1.3) |
C00044491
|
| 50846 | DHH, GDXYM, HHG-3, SRXY7 | desert hedgehog |
C00044491
|
| 1723 | DHODH, DHOdehase, POADS, URA1 | dihydroorotate dehydrogenase (quinone) (EC:1.3.5.2) |
C00044491
|
| 115817 | DHRS1, SDR19C1 | dehydrogenase/reductase (SDR family) member 1 |
C00044491
|
| 10202 | DHRS2, HEP27, SDR25C1 | dehydrogenase/reductase (SDR family) member 2 |
C00044491
|
| 9249 | DHRS3, DD83.1, RDH17, Rsdr1, SDR1, SDR16C1, retSDR1 | dehydrogenase/reductase (SDR family) member 3 (EC:1.1.1.300) |
C00044491
|
| 10170 | DHRS9, 3ALPHA-HSD, RDH-TBE, RDH15, RDHL, RDHTBE, RETSDR8, SDR9C4 | dehydrogenase/reductase (SDR family) member 9 |
C00044491
|
| 22907 | DHX30, DDX30, RETCOR | DEAH (Asp-Glu-Ala-His) box helicase 30 (EC:3.6.4.13) |
C00044491
|
| 56919 | DHX33, DDX33 | DEAH (Asp-Glu-Ala-His) box polypeptide 33 (EC:3.6.4.13) |
C00044491
|
| 1729 | DIAPH1, DFNA1, DIA1, DRF1, LFHL1, hDIA1 | diaphanous-related formin 1 |
C00044491
|
| 1730 | DIAPH2, DIA, DIA2, DRF2, POF, POF2 | diaphanous-related formin 2 |
C00044491
|
| 81624 | DIAPH3, AN, AUNA1, DIA2, DRF3, NSDAN, diap3, mDia2 | diaphanous-related formin 3 |
C00044491
|
| 23405 | DICER1, DCR1, Dicer, HERNA, MNG1, RMSE2 | dicer 1, ribonuclease type III (EC:3.1.26.-) |
C00044491
|
| 27042 | DIEXF, C1orf107, DEF, DJ434O14.5, UTP25 | digestive organ expansion factor homolog (zebrafish) |
C00044491
|
| 22982 | DIP2C, KIAA0934 | DIP2 disco-interacting protein 2 homolog C (Drosophila) |
C00044491
|
| 84925 | DIRC2, RCC4 | disrupted in renal carcinoma 2 |
C00044491
|
| 115752 | DIS3L, DIS3L1 | DIS3 mitotic control homolog (S. cerevisiae)-like (EC:3.1.13.-) |
C00044491
|
| 1737 | DLAT, DLTA, PDC-E2, PDCE2 | dihydrolipoamide S-acetyltransferase (EC:2.3.1.12) |
C00044491
|
| 1739 | DLG1, DLGH1, SAP-97, SAP97, dJ1061C18.1.1, hdlg | discs, large homolog 1 (Drosophila) |
C00044491
|
| 9231 | DLG5, LP-DLG, P-DLG5, PDLG | discs, large homolog 5 (Drosophila) |
C00044491
|
| 649446 | DLGAP1-AS1, HsT914 | DLGAP1 antisense RNA 1 |
C00044491
|
| 22839 | DLGAP4, DAP4, DLP4, RP5-977B1.6, SAPAP4 | discs, large (Drosophila) homolog-associated protein 4 |
C00044491
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00044491
|
| 1747 | DLX3, AI4, TDO | distal-less homeobox 3 |
C00044491
|
| 127343 | DMBX1, MBX, OTX3, PAXB | diencephalon/mesencephalon homeobox 1 |
C00044491
|
| 1756 | DMD, BMD, CMD3B, DXS142, DXS164, DXS206, DXS230, DXS239, DXS268, DXS269, DXS270, DXS272 | dystrophin |
C00044491
|
| 29958 | DMGDH, DMGDHD, ME2GLYDH | dimethylglycine dehydrogenase (EC:1.5.8.4) |
C00044491
|
| 1760 | DMPK, DM, DM1, DM1PK, DMK, MDPK, MT-PK | dystrophia myotonica-protein kinase (EC:2.7.11.1) |
C00044491
|
| 1657 | DMXL1 | Dmx-like 1 |
C00044491
|
| 23312 | DMXL2, RC3 | Dmx-like 2 |
C00044491
|
| 55172 | DNAAF2, C14orf104, CILD10, KTU, PF13 | dynein, axonemal, assembly factor 2 |
C00044491
|
| 3301 | DNAJA1, DJ-2, DjA1, HDJ2, HSDJ, HSJ2, HSPF4, NEDD7, hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 |
C00044491
|
| 55466 | DNAJA4, MST104, MSTP104, PRO1472 | DnaJ (Hsp40) homolog, subfamily A, member 4 |
C00044491
|
| 3337 | DNAJB1, HSPF1, Hdj1, Hsp40, RSPH16B, Sis1 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
C00044491
|
| 11080 | DNAJB4, DNAJW, DjB4, HLJ1 | DnaJ (Hsp40) homolog, subfamily B, member 4 |
C00044491
|
| 25822 | DNAJB5, Hsc40 | DnaJ (Hsp40) homolog, subfamily B, member 5 |
C00044491
|
| 10049 | DNAJB6, DJ4, DnaJ, HHDJ1, HSJ-2, HSJ2, LGMD1E, MRJ, MSJ-1 | DnaJ (Hsp40) homolog, subfamily B, member 6 |
C00044491
|
| 55735 | DNAJC11, dJ126A5.1 | DnaJ (Hsp40) homolog, subfamily C, member 11 |
C00044491
|
| 56521 | DNAJC12, JDP1 | DnaJ (Hsp40) homolog, subfamily C, member 12 |
C00044491
|
| 23317 | DNAJC13, RME8 | DnaJ (Hsp40) homolog, subfamily C, member 13 |
C00044491
|
| 134218 | DNAJC21, DNAJA5, GS3, JJJ1 | DnaJ (Hsp40) homolog, subfamily C, member 21 |
C00044491
|
| 7266 | DNAJC7, DJ11, DJC7, TPR2, TTC2 | DnaJ (Hsp40) homolog, subfamily C, member 7 |
C00044491
|
| 23234 | DNAJC9, HDJC9, JDD1, SB73 | DnaJ (Hsp40) homolog, subfamily C, member 9 |
C00044491
|
| 83544 | DNAL1, C14orf168, CILD16 | dynein, axonemal, light chain 1 |
C00044491
|
| 1773 | DNASE1, DNL1, DRNI | deoxyribonuclease I (EC:3.1.21.1) |
C00044491
|
| 1776 | DNASE1L3, DHP2, DNAS1L3, LSD, SLEB16 | deoxyribonuclease I-like 3 (EC:3.1.21.-) |
C00044491
|
| 23268 | DNMBP, ARHGEF36, TUBA | dynamin binding protein |
C00044491
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00044491
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00044491
|
| 80005 | DOCK5 | dedicator of cytokinesis 5 |
C00044491
|
| 85440 | DOCK7, ZIR2 | dedicator of cytokinesis 7 |
C00044491
|
| 220164 | DOK6, DOK5L, HsT3226 | docking protein 6 |
C00044491
|
| 9980 | DOPEY2, 21orf5, C21orf5 | dopey family member 2 |
C00044491
|
| 5977 | DPF2, REQ, UBID4, ubi-d4 | D4, zinc and double PHD fingers family 2 |
C00044491
|
| 51611 | DPH5, CGI-30, HSPC143, NPD015 | diphthamide biosynthesis 5 (EC:2.1.1.98) |
C00044491
|
| 8818 | DPM2, CDG1U | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (EC:2.4.1.83) |
C00044491
|
| 10072 | DPP3, DPPIII | dipeptidyl-peptidase 3 (EC:3.4.14.4) |
C00044491
|
| 1803 | DPP4, ADABP, ADCP2, CD26, DPPIV, TP103 | dipeptidyl-peptidase 4 (EC:3.4.14.5) |
C00044491
|
| 91039 | DPP9, DP9, DPLP9, DPRP-2, DPRP2 | dipeptidyl-peptidase 9 (EC:3.4.14.5) |
C00044491
|
| 147991 | DPY19L3 | dpy-19-like 3 (C. elegans) |
C00044491
|
| 1808 | DPYSL2, CRMP-2, CRMP2, DHPRP2, DRP-2, DRP2, N2A3, ULIP-2, ULIP2 | dihydropyrimidinase-like 2 |
C00044491
|
| 10570 | DPYSL4, CRMP3, DRP-4, ULIP4 | dihydropyrimidinase-like 4 |
C00044491
|
| 55332 | DRAM1, DRAM | DNA-damage regulated autophagy modulator 1 |
C00044491
|
| 128338 | DRAM2, PRO180, RP5-1180E21.1, TMEM77, WWFQ154 | DNA-damage regulated autophagy modulator 2 |
C00044491
|
| 10589 | DRAP1, NC2-alpha | DR1-associated protein 1 (negative cofactor 2 alpha) |
C00044491
|
| 1823 | DSC1, CDHF1, DG2/DG3 | desmocollin 1 |
C00044491
|
| 79075 | DSCC1, DCC1 | DNA replication and sister chromatid cohesion 1 |
C00044491
|
| 29940 | DSE, DSEP, DSEPI, SART-2, SART2 | dermatan sulfate epimerase (EC:5.1.3.19) |
C00044491
|
| 79980 | DSN1, C20orf172, KNL3, MIS13, dJ469A13.2, hKNL-3 | DSN1, MIS12 kinetochore complex component |
C00044491
|
| 667 | DST, BP240, BPA, BPAG1, CATX-15, CATX15, D6S1101, DMH, DT, EBSB2, HSAN6, MACF2 | dystonin |
C00044491
|
| 51514 | DTL, CDT2, DCAF2, L2DTL, RAMP | denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
C00044491
|
| 1838 | DTNB | dystrobrevin, beta |
C00044491
|
| 56986 | DTWD1 | DTW domain containing 1 |
C00044491
|
| 151636 | DTX3L, BBAP | deltex 3-like (Drosophila) |
C00044491
|
| 23220 | DTX4, RNF155 | deltex homolog 4 (Drosophila) |
C00044491
|
| 1841 | DTYMK, CDC8, PP3731, TMPK, TYMK | deoxythymidylate kinase (thymidylate kinase) (EC:2.7.4.9) |
C00044491
|
| 50506 | DUOX2, LNOX2, NOXEF2, P138-TOX, TDH6, THOX2 | dual oxidase 2 (EC:1.6.3.1) |
C00044491
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00044491
|
| 11221 | DUSP10, MKP-5, MKP5 | dual specificity phosphatase 10 (EC:3.1.3.16 3.1.3.48) |
C00044491
|
| 80824 | DUSP16, MKP-7, MKP7 | dual specificity phosphatase 16 (EC:3.1.3.16 3.1.3.48) |
C00044491
|
| 54808 | DYM, DMC, SMC | dymeclin |
C00044491
|
| 83658 | DYNLRB1, BITH, BLP, DNCL2A, DNLC2A, ROBLD1 | dynein, light chain, roadblock-type 1 |
C00044491
|
| 6993 | DYNLT1, CW-1, TCTEL1, tctex-1 | dynein, light chain, Tctex-type 1 |
C00044491
|
| 8445 | DYRK2 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (EC:2.7.12.1) |
C00044491
|
| 8444 | DYRK3, DYRK5, RED, REDK, hYAK3-2 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 (EC:2.7.12.1) |
C00044491
|
| 8291 | DYSF, FER1L1, LGMD2B, MMD1 | dysferlin |
C00044491
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00044491
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00044491
|
| 1875 | E2F5, E2F-5 | E2F transcription factor 5, p130-binding |
C00044491
|
| 144455 | E2F7 | E2F transcription factor 7 |
C00044491
|
| 55837 | EAPP, BM036, C14orf11 | E2F-associated phosphoprotein |
C00044491
|
| 11319 | ECD, GCR2, HSGT1 | ecdysoneless homolog (Drosophila) |
C00044491
|
| 1889 | ECE1, ECE | endothelin converting enzyme 1 (EC:3.4.24.71) |
C00044491
|
| 9427 | ECEL1, DA5D, DINE, ECEX, XCE | endothelin converting enzyme-like 1 |
C00044491
|
| 55268 | ECHDC2 | enoyl CoA hydratase domain containing 2 |
C00044491
|
| 10455 | ECI2, ACBD2, DRS-1, DRS1, HCA88, PECI, dJ1013A10.3 | enoyl-CoA delta isomerase 2 (EC:5.3.3.8) |
C00044491
|
| 641700 | ECSCR, ARIA, ECSM2 | endothelial cell surface expressed chemotaxis and apoptosis regulator |
C00044491
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00044491
|
| 80267 | EDEM3, C1orf22 | ER degradation enhancer, mannosidase alpha-like 3 (EC:3.2.1.113) |
C00044491
|
| 1910 | EDNRB, ABCDS, ET-B, ET-BR, ETB, ETBR, ETRB, HSCR, HSCR2, WS4A | endothelin receptor type B |
C00044491
|
| 8411 | EEA1, MST105, MSTP105, ZFYVE2 | early endosome antigen 1 |
C00044491
|
| 8726 | EED, HEED, WAIT1 | embryonic ectoderm development |
C00044491
|
| 1936 | EEF1D, EF-1D, EF1D, FP1047 | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
C00044491
|
| 9521 | EEF1E1, AIMP3, P18 | eukaryotic translation elongation factor 1 epsilon 1 |
C00044491
|
| 29904 | EEF2K, HSU93850, eEF-2K | eukaryotic elongation factor-2 kinase (EC:2.7.11.20) |
C00044491
|
| 80820 | EEPD1, HSPC107 | endonuclease/exonuclease/phosphatase family domain containing 1 |
C00044491
|
| 100130771 | EFCAB10 | EF-hand calcium binding domain 10 |
C00044491
|
| 9813 | EFCAB14, KIAA0494, RP11-8J9.3 | EF-hand calcium binding domain 14 |
C00044491
|
| 79825 | EFCC1, C3orf73, CCDC48 | EF-hand and coiled-coil domain containing 1 |
C00044491
|
| 30008 | EFEMP2, ARCL1B, FBLN4, MBP1, UPH1 | EGF containing fibulin-like extracellular matrix protein 2 |
C00044491
|
| 114327 | EFHC1, dJ304B14.2 | EF-hand domain (C-terminal) containing 1 |
C00044491
|
| 1942 | EFNA1, B61, ECKLG, EFL1, EPLG1, LERK-1, LERK1, TNFAIP4 | ephrin-A1 |
C00044491
|
| 1943 | EFNA2, ELF-1, EPLG6, HEK7-L, LERK-6, LERK6 | ephrin-A2 |
C00044491
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00044491
|
| 23167 | EFR3A | EFR3 homolog A (S. cerevisiae) |
C00044491
|
| 79631 | EFTUD1, FAM42A, HsT19294, RIA1 | elongation factor Tu GTP binding domain containing 1 |
C00044491
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00044491
|
| 54583 | EGLN1, C1orf12, ECYT3, HIF-PH2, HIFPH2, HPH-2, HPH2, PHD2, SM20, ZMYND6 | egl-9 family hypoxia-inducible factor 1 (EC:1.14.11.29) |
C00044491
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00044491
|
| 23301 | EHBP1, HPC12, NACSIN | EH domain binding protein 1 |
C00044491
|
| 9538 | EI24, EPG4, PIG8, TP53I8 | etoposide induced 2.4 |
C00044491
|
| 10209 | EIF1, A121, EIF-1, EIF1A, ISO1, SUI1 | eukaryotic translation initiation factor 1 |
C00044491
|
| 83939 | EIF2A, EIF-2A, MST089, MSTP004, MSTP089 | eukaryotic translation initiation factor 2A, 65kDa |
C00044491
|
| 5610 | EIF2AK2, EIF2AK1, PKR, PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 (EC:2.7.11.1 2.7.10.2) |
C00044491
|
| 440275 | EIF2AK4, GCN2 | eukaryotic translation initiation factor 2 alpha kinase 4 (EC:2.7.11.1) |
C00044491
|
| 8661 | EIF3A, EIF3, EIF3S10, P167, TIF32, eIF3-p170, eIF3-theta, p180, p185 | eukaryotic translation initiation factor 3, subunit A |
C00044491
|
| 3646 | EIF3E, EIF3-P48, EIF3S6, INT6, eIF3-p46 | eukaryotic translation initiation factor 3, subunit E |
C00044491
|
| 8665 | EIF3F, EIF3S5, eIF3-p47 | eukaryotic translation initiation factor 3, subunit F (EC:3.4.19.12) |
C00044491
|
| 1973 | EIF4A1, DDX2A, EIF-4A, EIF4A, eIF-4A-I, eIF4A-I | eukaryotic translation initiation factor 4A1 (EC:3.6.4.13) |
C00044491
|
| 1975 | EIF4B, EIF-4B, PRO1843 | eukaryotic translation initiation factor 4B |
C00044491
|
| 1981 | EIF4G1, EIF-4G1, EIF4F, EIF4G, EIF4GI, P220, PARK18 | eukaryotic translation initiation factor 4 gamma, 1 |
C00044491
|
| 8672 | EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII | eukaryotic translation initiation factor 4 gamma, 3 |
C00044491
|
| 1984 | EIF5A, EIF-5A, EIF5A1, eIF5AI | eukaryotic translation initiation factor 5A |
C00044491
|
| 60528 | ELAC2, COXPD17, ELC2, HPC2 | elaC ribonuclease Z 2 (EC:3.1.26.11) |
C00044491
|
| 1994 | ELAVL1, ELAV1, HUR, Hua, MelG | ELAV like RNA binding protein 1 |
C00044491
|
| 1997 | ELF1 | E74-like factor 1 (ets domain transcription factor) |
C00044491
|
| 1999 | ELF3, EPR-1, ERT, ESE-1, ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
C00044491
|
| 2000 | ELF4, ELFR, MEF | E74-like factor 4 (ets domain transcription factor) |
C00044491
|
| 2004 | ELK3, ERP, NET, SAP2 | ELK3, ETS-domain protein (SRF accessory protein 2) |
C00044491
|
| 22936 | ELL2 | elongation factor, RNA polymerase II, 2 |
C00044491
|
| 9844 | ELMO1, CED-12, CED12, ELMO-1 | engulfment and cell motility 1 |
C00044491
|
| 63916 | ELMO2, CED-12, CED12, ELMO-2 | engulfment and cell motility 2 |
C00044491
|
| 255520 | ELMOD2, 9830169G11Rik | ELMO/CED-12 domain containing 2 |
C00044491
|
| 84173 | ELMOD3, DFNB88, LST3, RBED1, RBM29 | ELMO/CED-12 domain containing 3 |
C00044491
|
| 64834 | ELOVL1, Ssc1 | ELOVL fatty acid elongase 1 (EC:2.3.1.199) |
C00044491
|
| 54898 | ELOVL2, SSC2 | ELOVL fatty acid elongase 2 (EC:2.3.1.199) |
C00044491
|
| 60481 | ELOVL5, HELO1, dJ483K16.1 | ELOVL fatty acid elongase 5 (EC:2.3.1.199) |
C00044491
|
| 79071 | ELOVL6, FACE, FAE, LCE | ELOVL fatty acid elongase 6 (EC:2.3.1.199) |
C00044491
|
| 79993 | ELOVL7 | ELOVL fatty acid elongase 7 (EC:2.3.1.199) |
C00044491
|
| 55250 | ELP2, SHINC-2, STATIP1, StIP | elongator acetyltransferase complex subunit 2 |
C00044491
|
| 55140 | ELP3, KAT9 | elongator acetyltransferase complex subunit 3 (EC:2.3.1.48) |
C00044491
|
| 54859 | ELP6, C3orf75, TMEM103 | elongator acetyltransferase complex subunit 6 |
C00044491
|
| 55831 | EMC3, POB, TMEM111 | ER membrane protein complex subunit 3 |
C00044491
|
| 146956 | EME1, MMS4L, SLX2A | essential meiotic structure-specific endonuclease 1 |
C00044491
|
| 197342 | EME2, SLX2B, gs125 | essential meiotic structure-specific endonuclease subunit 2 |
C00044491
|
| 256364 | EML3, ELP95 | echinoderm microtubule associated protein like 3 |
C00044491
|
| 27436 | EML4, C2orf2, ELP120, EMAP-4, EMAPL4, ROPP120 | echinoderm microtubule associated protein like 4 |
C00044491
|
| 2013 | EMP2, XMP | epithelial membrane protein 2 |
C00044491
|
| 2019 | EN1 | engrailed homeobox 1 |
C00044491
|
| 55740 | ENAH, ENA, MENA, NDPP1 | enabled homolog (Drosophila) |
C00044491
|
| 8507 | ENC1, CCL28, ENC-1, KLHL35, KLHL37, NRPB, PIG10, TP53I10 | ectodermal-neural cortex 1 (with BTB domain) |
C00044491
|
| 23052 | ENDOD1 | endonuclease domain containing 1 |
C00044491
|
| 2023 | ENO1, ENO1L1, MPB1, NNE, PPH | enolase 1, (alpha) (EC:4.2.1.11) |
C00044491
|
| 2028 | ENPEP, APA, CD249, gp160 | glutamyl aminopeptidase (aminopeptidase A) (EC:3.4.11.7) |
C00044491
|
| 5169 | ENPP3, B10, CD203c, NPP3, PD-IBETA, PDNP3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 (EC:3.1.4.1 3.6.1.9) |
C00044491
|
| 2029 | ENSA, ARPP-19e | endosulfine alpha |
C00044491
|
| 957 | ENTPD5, CD39L4, NTPDase-5, PCPH | ectonucleoside triphosphate diphosphohydrolase 5 (EC:3.6.1.6 3.6.1.42) |
C00044491
|
| 955 | ENTPD6, CD39L2, IL-6SAG, IL6ST2, NTPDase-6, dJ738P15.3 | ectonucleoside triphosphate diphosphohydrolase 6 (putative) (EC:3.6.1.6) |
C00044491
|
| 2035 | EPB41, 4.1R, EL1, HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
C00044491
|
| 114915 | EPB41L4A-AS1, C5orf26, NCRNA00219, TIGA1 | EPB41L4A antisense RNA 1 |
C00044491
|
| 54566 | EPB41L4B, CG1, EHM2 | erythrocyte membrane protein band 4.1 like 4B |
C00044491
|
| 57669 | EPB41L5, BE37, YMO1 | erythrocyte membrane protein band 4.1 like 5 |
C00044491
|
| 2050 | EPHB4, HTK, MYK1, TYRO11 | EPH receptor B4 (EC:2.7.10.1) |
C00044491
|
| 2052 | EPHX1, EPHX, EPOX, HYL1, MEH | epoxide hydrolase 1, microsomal (xenobiotic) (EC:3.3.2.9) |
C00044491
|
| 9852 | EPM2AIP1 | EPM2A (laforin) interacting protein 1 |
C00044491
|
| 2059 | EPS8 | epidermal growth factor receptor pathway substrate 8 |
C00044491
|
| 64787 | EPS8L2, EPS8R2 | EPS8-like 2 |
C00044491
|
| 2064 | ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NGL, TKR1 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (EC:2.7.10.1) |
C00044491
|
| 55914 | ERBB2IP, ERBIN, LAP2 | erbb2 interacting protein |
C00044491
|
| 2065 | ERBB3, ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 (EC:2.7.10.1) |
C00044491
|
| 2071 | ERCC3, BTF2, GTF2H, RAD25, TFIIH, XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (EC:3.6.4.12) |
C00044491
|
| 54821 | ERCC6L, PICH, RAD26L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like (EC:3.6.4.12) |
C00044491
|
| 51290 | ERGIC2, Erv41, PTX1, cd002 | ERGIC and golgi 2 |
C00044491
|
| 90459 | ERI1, 3'HEXO, HEXO, THEX1 | exoribonuclease 1 |
C00044491
|
| 112479 | ERI2, EXOD1 | ERI1 exoribonuclease family member 2 (EC:3.1.-.-) |
C00044491
|
| 2081 | ERN1, IRE1, IRE1P, IRE1a, hIRE1p | endoplasmic reticulum to nucleus signaling 1 (EC:2.7.11.1) |
C00044491
|
| 30001 | ERO1L, ERO1-alpha, ERO1A, ERO1LA | ERO1-like (S. cerevisiae) |
C00044491
|
| 54206 | ERRFI1, GENE-33, MIG-6, MIG6, RALT | ERBB receptor feedback inhibitor 1 |
C00044491
|
| 157570 | ESCO2, 2410004I17Rik, EFO2, RBS | establishment of sister chromatid cohesion N-acetyltransferase 2 (EC:2.3.1.-) |
C00044491
|
| 9700 | ESPL1, ESP1, SEPA | extra spindle pole bodies homolog 1 (S. cerevisiae) (EC:3.4.22.49) |
C00044491
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00044491
|
| 23344 | ESYT1, FAM62A, MBC2 | extended synaptotagmin-like protein 1 |
C00044491
|
| 2109 | ETFB, MADD | electron-transfer-flavoprotein, beta polypeptide |
C00044491
|
| 55500 | ETNK1, EKI, EKI_1, EKI1, Nbla10396 | ethanolamine kinase 1 (EC:2.7.1.82) |
C00044491
|
| 64850 | ETNPPL, AGXT2L1 | ethanolamine-phosphate phospho-lyase (EC:4.2.3.2) |
C00044491
|
| 2114 | ETS2, ETS2IT1 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
C00044491
|
| 2119 | ETV5, ERM | ets variant 5 |
C00044491
|
| 2120 | ETV6, TEL, TEL/ABL | ets variant 6 |
C00044491
|
| 84141 | EVA1A, FAM176A, TMEM166 | eva-1 homolog A (C. elegans) |
C00044491
|
| 7813 | EVI5, NB4S | ecotropic viral integration site 5 |
C00044491
|
| 115704 | EVI5L | ecotropic viral integration site 5-like |
C00044491
|
| 55218 | EXD2, C14orf114, EXDL2 | exonuclease 3'-5' domain containing 2 |
C00044491
|
| 9156 | EXO1, HEX1, hExoI | exonuclease 1 |
C00044491
|
| 64789 | EXO5, C1orf176, DEM1, Exo_V, hExo5 | exonuclease 5 |
C00044491
|
| 55763 | EXOC1, BM-102, SEC3, SEC3L1, SEC3P | exocyst complex component 1 |
C00044491
|
| 10640 | EXOC5, HSEC10, PRO1912, SEC10, SEC10L1, SEC10P | exocyst complex component 5 |
C00044491
|
| 23233 | EXOC6B, SEC15B, SEC15L2 | exocyst complex component 6B |
C00044491
|
| 2131 | EXT1, EXT, LGCR, LGS, TRPS2, TTV | exostosin glycosyltransferase 1 (EC:2.4.1.225 2.4.1.224) |
C00044491
|
| 2132 | EXT2, SOTV | exostosin glycosyltransferase 2 (EC:2.4.1.225 2.4.1.224) |
C00044491
|
| 2135 | EXTL2, EXTR2 | exostosin-like glycosyltransferase 2 (EC:2.4.1.223) |
C00044491
|
| 2140 | EYA3 | eyes absent homolog 3 (Drosophila) (EC:3.1.3.48) |
C00044491
|
| 2146 | EZH2, ENX-1, ENX1, EZH1, EZH2b, KMT6, KMT6A, WVS, WVS2 | enhancer of zeste homolog 2 (Drosophila) (EC:2.1.1.43) |
C00044491
|
| 7430 | EZR, CVIL, CVL, VIL2 | ezrin |
C00044491
|
| 9002 | F2RL3, PAR4 | coagulation factor II (thrombin) receptor-like 3 |
C00044491
|
| 2166 | FAAH, FAAH-1, PSAB | fatty acid amide hydrolase (EC:3.5.1.99) |
C00044491
|
| 2168 | FABP1, FABPL, L-FABP | fatty acid binding protein 1, liver |
C00044491
|
| 2167 | FABP4, A-FABP, AFABP, ALBP, aP2 | fatty acid binding protein 4, adipocyte |
C00044491
|
| 2171 | FABP5, E-FABP, EFABP, KFABP, PA-FABP, PAFABP | fatty acid binding protein 5 (psoriasis-associated) |
C00044491
|
| 8772 | FADD, MORT1 | Fas (TNFRSF6)-associated via death domain |
C00044491
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00044491
|
| 9415 | FADS2, D6D, DES6, FADSD6, LLCDL2, SLL0262, TU13 | fatty acid desaturase 2 (EC:1.14.19.-) |
C00044491
|
| 3995 | FADS3, CYB5RP, LLCDL3 | fatty acid desaturase 3 (EC:1.14.19.-) |
C00044491
|
| 81889 | FAHD1, C16orf36, YISKL | fumarylacetoacetate hydrolase domain containing 1 (EC:3.7.1.5) |
C00044491
|
| 55179 | FAIM, FAIM1 | Fas apoptotic inhibitory molecule |
C00044491
|
| 83640 | FAM103A1, C15orf18, HsT19360, RAM | family with sequence similarity 103, member A1 |
C00044491
|
| 83641 | FAM107B, C10orf45 | family with sequence similarity 107, member B |
C00044491
|
| 63901 | FAM111A, GCLEB, KCS2 | family with sequence similarity 111, member A |
C00044491
|
| 92689 | FAM114A1, Noxp20 | family with sequence similarity 114, member A1 |
C00044491
|
| 10827 | FAM114A2, 133K02, C5orf3 | family with sequence similarity 114, member A2 |
C00044491
|
| 285966 | FAM115C, FAM139A | family with sequence similarity 115, member C |
C00044491
|
| 55007 | FAM118A, C22orf8 | family with sequence similarity 118, member A |
C00044491
|
| 79607 | FAM118B | family with sequence similarity 118, member B |
C00044491
|
| 23196 | FAM120A, C9orf10, DNAPTP1, DNAPTP5, OSSA | family with sequence similarity 120A |
C00044491
|
| 116224 | FAM122A, C9orf42 | family with sequence similarity 122A |
C00044491
|
| 159090 | FAM122B, SPACIA2 | family with sequence similarity 122B |
C00044491
|
| 84668 | FAM126A, DRCTNNB1A, HCC, HLD5, HYCC1 | family with sequence similarity 126, member A |
C00044491
|
| 162427 | FAM134C | family with sequence similarity 134, member C |
C00044491
|
| 57579 | FAM135A, KIAA1411 | family with sequence similarity 135, member A |
C00044491
|
| 25854 | FAM149A, MSTP119 | family with sequence similarity 149, member A |
C00044491
|
| 389658 | FAM150A, UNQ9433 | family with sequence similarity 150, member A |
C00044491
|
| 64760 | FAM160B2, RAI16 | family with sequence similarity 160, member B2 |
C00044491
|
| 84140 | FAM161A, RP28 | family with sequence similarity 161, member A |
C00044491
|
| 221061 | FAM171A1, C10orf38 | family with sequence similarity 171, member A1 |
C00044491
|
| 284069 | FAM171A2 | family with sequence similarity 171, member A2 |
C00044491
|
| 55719 | FAM178A, C10orf6 | family with sequence similarity 178, member A |
C00044491
|
| 10712 | FAM189B, C1orf2, COTE1 | family with sequence similarity 189, member B |
C00044491
|
| 51313 | FAM198B, AD036, C4orf18 | family with sequence similarity 198, member B |
C00044491
|
| 23272 | FAM208A, C3orf63, RAP140, se89-1 | family with sequence similarity 208, member A |
C00044491
|
| 54757 | FAM20A, AIGFS, FP2747 | family with sequence similarity 20, member A |
C00044491
|
| 116151 | FAM210B, 5A3, C20orf108, dJ1167H4.1 | family with sequence similarity 210, member B |
C00044491
|
| 55924 | FAM212B, C1orf183 | family with sequence similarity 212, member B |
C00044491
|
| 23591 | FAM215A, APR-2, C17orf88, LINC00530 | family with sequence similarity 215, member A (non-protein coding) |
C00044491
|
| 84792 | FAM220A, C7orf70, SIPAR | family with sequence similarity 220, member A |
C00044491
|
| 55731 | FAM222B, C17orf63 | family with sequence similarity 222, member B |
C00044491
|
| 196951 | FAM227B, C15orf33 | family with sequence similarity 227, member B |
C00044491
|
| 284123 | FAM27L | family with sequence similarity 27-like |
C00044491
|
| 366403 |
C00044491
|
||
| 54537 | FAM35A, FAM35A1, bA163M19.1 | family with sequence similarity 35, member A |
C00044491
|
| 55603 | FAM46A, C6orf37, XTP11 | family with sequence similarity 46, member A |
C00044491
|
| 54855 | FAM46C | family with sequence similarity 46, member C |
C00044491
|
| 9130 | FAM50A, 9F, DXS9928E, HXC-26, HXC26, XAP5 | family with sequence similarity 50, member A |
C00044491
|
| 51307 | FAM53C, C5orf6 | family with sequence similarity 53, member C |
C00044491
|
| 83723 | FAM57B | family with sequence similarity 57, member B |
C00044491
|
| 58516 | FAM60A, C12orf14, TERA | family with sequence similarity 60, member A |
C00044491
|
| 79567 | FAM65A | family with sequence similarity 65, member A |
C00044491
|
| 374986 | FAM73A | family with sequence similarity 73, member A |
C00044491
|
| 199870 | FAM76A | family with sequence similarity 76, member A |
C00044491
|
| 81610 | FAM83D, C20orf129, CHICA, dJ616B8.3 | family with sequence similarity 83, member D |
C00044491
|
| 644815 | FAM83G | family with sequence similarity 83, member G |
C00044491
|
| 286077 | FAM83H, AI3 | family with sequence similarity 83, member H |
C00044491
|
| 85002 | FAM86B1 | family with sequence similarity 86, member B1 |
C00044491
|
| 692099 | FAM86DP, FAM86D | family with sequence similarity 86, member D, pseudogene |
C00044491
|
| 22909 | FAN1, KIAA1018, KMIN, MTMR15 | FANCD2/FANCI-associated nuclease 1 (EC:3.1.4.1) |
C00044491
|
| 2175 | FANCA, FA, FA-H, FA1, FAA, FACA, FAH, FANCH | Fanconi anemia, complementation group A |
C00044491
|
| 2187 | FANCB, FA2, FAAP90, FAAP95, FAB, FACB | Fanconi anemia, complementation group B |
C00044491
|
| 2177 | FANCD2, FA-D2, FA4, FACD, FAD, FAD2, FANCD | Fanconi anemia, complementation group D2 |
C00044491
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00044491
|
| 55120 | FANCL, FAAP43, PHF9, POG | Fanconi anemia, complementation group L |
C00044491
|
| 57697 | FANCM, FAAP250, KIAA1596 | Fanconi anemia, complementation group M (EC:3.6.4.13) |
C00044491
|
| 10160 | FARP1, CDEP, PLEKHC2, PPP1R75 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
C00044491
|
| 9855 | FARP2, FIR, FRG, PLEKHC3 | FERM, RhoGEF and pleckstrin domain protein 2 |
C00044491
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00044491
|
| 2194 | FASN, FAS, OA-519, SDR27X1 | fatty acid synthase (EC:2.3.1.85 2.3.1.38 2.3.1.39 2.3.1.41 3.1.2.14 4.2.1.59 1.1.1.100 1.3.1.39) |
C00044491
|
| 79675 | FASTKD1 | FAST kinase domains 1 |
C00044491
|
| 22868 | FASTKD2, KIAA0971 | FAST kinase domains 2 |
C00044491
|
| 79072 | FASTKD3 | FAST kinase domains 3 |
C00044491
|
| 2195 | FAT1, CDHF7, CDHR8, FAT, ME5, hFat1 | FAT atypical cadherin 1 |
C00044491
|
| 10826 | FAXDC2, C5orf4 | fatty acid hydroxylase domain containing 2 |
C00044491
|
| 54751 | FBLIM1, CAL, FBLP-1, FBLP1 | filamin binding LIM protein 1 |
C00044491
|
| 2192 | FBLN1, FBLN, FIBL1 | fibulin 1 |
C00044491
|
| 2201 | FBN2, CCA, DA9 | fibrillin 2 |
C00044491
|
| 64839 | FBXL17, FBXO13, Fbl17, Fbx13 | F-box and leucine-rich repeat protein 17 |
C00044491
|
| 84961 | FBXL20, Fbl2, Fbl20 | F-box and leucine-rich repeat protein 20 |
C00044491
|
| 26223 | FBXL21, FBL3B, FBXL3B, FBXL3P, Fbl21 | F-box and leucine-rich repeat protein 21 (gene/pseudogene) |
C00044491
|
| 84893 | FBXO18, FBH1, Fbx18 | F-box protein, helicase, 18 (EC:3.6.4.12) |
C00044491
|
| 26263 | FBXO22, FBX22, FISTC1 | F-box protein 22 |
C00044491
|
| 23219 | FBXO28, CENP-30, Fbx28 | F-box protein 28 |
C00044491
|
| 26273 | FBXO3, FBA, FBX3 | F-box protein 3 |
C00044491
|
| 84085 | FBXO30, Fbx30 | F-box protein 30 |
C00044491
|
| 254170 | FBXO33, BMND12, Fbx33, c14_5247 | F-box protein 33 |
C00044491
|
| 55030 | FBXO34, Fbx34 | F-box protein 34 |
C00044491
|
| 130888 | FBXO36, Fbx36 | F-box protein 36 |
C00044491
|
| 286151 | FBXO43, EMI2, ERP1, FBX43 | F-box protein 43 |
C00044491
|
| 93611 | FBXO44, FBG3, FBX30, FBX6A, Fbx44, Fbxo6a | F-box protein 44 |
C00044491
|
| 200933 | FBXO45, Fbx45 | F-box protein 45 |
C00044491
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00044491
|
| 26270 | FBXO6, FBG2, FBS2, FBX6, Fbx6b | F-box protein 6 |
C00044491
|
| 25793 | FBXO7, FBX, FBX07, FBX7, PARK15, PKPS | F-box protein 7 |
C00044491
|
| 26268 | FBXO9, FBX9, NY-REN-57, VCIA1, dJ341E18.2 | F-box protein 9 |
C00044491
|
| 23291 | FBXW11, BTRC2, BTRCP2, FBW1B, FBXW1B, Fbw11, Hos | F-box and WD repeat domain containing 11 |
C00044491
|
| 55294 | FBXW7, AGO, CDC4, FBW6, FBW7, FBX30, FBXO30, FBXW6, SEL-10, SEL10, hAgo, hCdc4 | F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
C00044491
|
| 83953 | FCAMR, CD351, FCA/MR | Fc receptor, IgA, IgM, high affinity |
C00044491
|
| 2208 | FCER2, BLAST-2, CD23, CD23A, CLEC4J, FCE2, IGEBF | Fc fragment of IgE, low affinity II, receptor for (CD23) |
C00044491
|
| 115548 | FCHO2 | FCH domain only 2 |
C00044491
|
| 9873 | FCHSD2, NWK, SH3MD3 | FCH and double SH3 domains 2 |
C00044491
|
| 2220 | FCN2, EBP-37, FCNL, P35, ficolin-2 | ficolin (collagen/fibrinogen domain containing lectin) 2 |
C00044491
|
| 2224 | FDPS, FPPS, FPS | farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10) |
C00044491
|
| 2232 | FDXR, ADXR | ferredoxin reductase (EC:1.18.1.6) |
C00044491
|
| 56929 | FEM1C, EUROIMAGE686608, EUROIMAGE783647, FEM1A | fem-1 homolog c (C. elegans) |
C00044491
|
| 2237 | FEN1, FEN-1, MF1, RAD2 | flap structure-specific endonuclease 1 |
C00044491
|
| 26998 | FETUB, 16G2, Gugu, IRL685 | fetuin B |
C00044491
|
| 2256 | FGF11, FHF3 | fibroblast growth factor 11 |
C00044491
|
| 26291 | FGF21 | fibroblast growth factor 21 |
C00044491
|
| 2260 | FGFR1, BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | fibroblast growth factor receptor 1 (EC:2.7.10.1) |
C00044491
|
| 26127 | FGFR1OP2, HSPC123-like, WIT3.0 | FGFR1 oncogene partner 2 |
C00044491
|
| 2261 | FGFR3, ACH, CD333, CEK2, HSFGFR3EX, JTK4 | fibroblast growth factor receptor 3 (EC:2.7.10.1) |
C00044491
|
| 114827 | FHAD1 | forkhead-associated (FHA) phosphopeptide binding domain 1 |
C00044491
|
| 80206 | FHOD3, FHOS2, Formactin2 | formin homology 2 domain containing 3 |
C00044491
|
| 55137 | FIGN | fidgetin |
C00044491
|
| 63979 | FIGNL1 | fidgetin-like 1 |
C00044491
|
| 161247 | FITM1, FIT1 | fat storage-inducing transmembrane protein 1 |
C00044491
|
| 128486 | FITM2, C20orf142, Fit2, dJ881L22.2 | fat storage-inducing transmembrane protein 2 |
C00044491
|
| 24147 | FJX1 | four jointed box 1 (Drosophila) |
C00044491
|
| 2288 | FKBP4, FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | FK506 binding protein 4, 59kDa (EC:5.2.1.8) |
C00044491
|
| 79147 | FKRP, LGMD2I, MDC1C, MDDGA5, MDDGB5, MDDGC5 | fukutin related protein |
C00044491
|
| 201163 | FLCN, BHD, FLCL | folliculin |
C00044491
|
| 2317 | FLNB, ABP-278, ABP-280, AOI, FH1, FLN-B, FLN1L, LRS1, SCT, TABP, TAP | filamin B, beta |
C00044491
|
| 2318 | FLNC, ABP-280, ABP280A, ABPA, ABPL, FLN2, MFM5, MPD4 | filamin C, gamma |
C00044491
|
| 2319 | FLOT2, ECS-1, ECS1, ESA, ESA1, M17S1 | flotillin 2 |
C00044491
|
| 23767 | FLRT3, HH21 | fibronectin leucine rich transmembrane protein 3 |
C00044491
|
| 55640 | FLVCR2, C14orf58, CCT, EPV, FLVCRL14q, MFSD7C, PVHH | feline leukemia virus subgroup C cellular receptor family, member 2 |
C00044491
|
| 84256 | FLYWCH1 | FLYWCH-type zinc finger 1 |
C00044491
|
| 91010 | FMNL3, FHOD3, WBP-3, WBP3 | formin-like 3 |
C00044491
|
| 2329 | FMO4, FMO2 | flavin containing monooxygenase 4 (EC:1.14.13.8) |
C00044491
|
| 2335 | FN1, CIG, ED-B, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | fibronectin 1 |
C00044491
|
| 79672 | FN3KRP, FN3KL | fructosamine 3 kinase related protein |
C00044491
|
| 23048 | FNBP1, FBP17 | formin binding protein 1 |
C00044491
|
| 22862 | FNDC3A, FNDC3, HUGO, bA203I16.1, bA203I16.5 | fibronectin type III domain containing 3A |
C00044491
|
| 64778 | FNDC3B, FAD104, PRO4979, YVTM2421 | fibronectin type III domain containing 3B |
C00044491
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00044491
|
| 2354 | FOSB, AP-1, G0S3, GOS3, GOSB | FBJ murine osteosarcoma viral oncogene homolog B |
C00044491
|
| 2355 | FOSL2, FRA2 | FOS-like antigen 2 |
C00044491
|
| 3170 | FOXA2, HNF3B, TCF3B | forkhead box A2 |
C00044491
|
| 221937 | FOXK1, FOXK1L | forkhead box K1 |
C00044491
|
| 3607 | FOXK2, ILF, ILF-1, ILF1 | forkhead box K2 |
C00044491
|
| 2305 | FOXM1, FKHL16, FOXM1B, HFH-11, HFH11, HNF-3, INS-1, MPHOSPH2, MPP-2, MPP2, PIG29, TGT3, TRIDENT | forkhead box M1 |
C00044491
|
| 2308 | FOXO1, FKH1, FKHR, FOXO1A | forkhead box O1 |
C00044491
|
| 2309 | FOXO3, AF6q21, FKHRL1, FKHRL1P2, FOXO2, FOXO3A | forkhead box O3 |
C00044491
|
| 27086 | FOXP1, 12CC4, QRF1, hFKH1B | forkhead box P1 |
C00044491
|
| 94234 | FOXQ1, HFH1 | forkhead box Q1 |
C00044491
|
| 2359 | FPR3, FML2_HUMAN, FMLPY, FPRH1, FPRH2, FPRL2, RMLP-R-I | formyl peptide receptor 3 |
C00044491
|
| 55691 | FRMD4A, FRMD4, bA295P9.4 | FERM domain containing 4A |
C00044491
|
| 84978 | FRMD5 | FERM domain containing 5 |
C00044491
|
| 122786 | FRMD6, C14orf31, EX1, Willin, c14_5320 | FERM domain containing 6 |
C00044491
|
| 83786 | FRMD8 | FERM domain containing 8 |
C00044491
|
| 143162 | FRMPD2, PDZD5C, PDZK4, PDZK5C | FERM and PDZ domain containing 2 |
C00044491
|
| 23732 | FRRS1L, C9orf4, CG-6, CG6 | ferric-chelate reductase 1-like |
C00044491
|
| 10129 | FRY, 13CDNA73, 214K23.2, C13orf14, CG003, bA207N4.2, bA37E23.1 | furry homolog (Drosophila) |
C00044491
|
| 285527 | FRYL, KIAA0826 | FRY-like |
C00044491
|
| 10468 | FST, FS | follistatin |
C00044491
|
| 10841 | FTCD, LCHC1 | formimidoyltransferase cyclodeaminase (EC:2.1.2.5 4.3.1.4) |
C00044491
|
| 2495 | FTH1, FHC, FTH, FTHL6, PIG15, PLIF | ferritin, heavy polypeptide 1 (EC:1.16.3.1) |
C00044491
|
| 24140 | FTSJ1, CDLIV, MRX44, MRX9, SPB1, TRMT7 | FtsJ RNA methyltransferase homolog 1 (E. coli) (EC:2.1.1.205) |
C00044491
|
| 8880 | FUBP1, FBP, FUBP | far upstream element (FUSE) binding protein 1 |
C00044491
|
| 2517 | FUCA1, FUCA | fucosidase, alpha-L- 1, tissue (EC:3.2.1.51) |
C00044491
|
| 139341 | FUNDC1 | FUN14 domain containing 1 |
C00044491
|
| 5045 | FURIN, FUR, PACE, PCSK3, SPC1 | furin (paired basic amino acid cleaving enzyme) (EC:3.4.21.75) |
C00044491
|
| 2521 | FUS, ALS6, ETM4, FUS1, HNRNPP2, POMP75, TLS | fused in sarcoma |
C00044491
|
| 2528 | FUT6, FCT3A, FT1A, Fuc-TVI, FucT-VI | fucosyltransferase 6 (alpha (1,3) fucosyltransferase) (EC:2.4.1.65) |
C00044491
|
| 8087 | FXR1, FXR1P | fragile X mental retardation, autosomal homolog 1 |
C00044491
|
| 5348 | FXYD1, PLM | FXYD domain containing ion transport regulator 1 |
C00044491
|
| 486 | FXYD2, ATP1G1, HOMG2 | FXYD domain containing ion transport regulator 2 |
C00044491
|
| 79443 | FYCO1, CATC2, CTRCT18, RUFY3, ZFYVE7 | FYVE and coiled-coil domain containing 1 |
C00044491
|
| 2534 | FYN, SLK, SYN, p59-FYN | FYN oncogene related to SRC, FGR, YES (EC:2.7.10.2) |
C00044491
|
| 8322 | FZD4, CD344, EVR1, FEVR, FZD4S, Fz-4, Fz4, FzE4, GPCR, hFz4 | frizzled family receptor 4 |
C00044491
|
| 7855 | FZD5, C2orf31, HFZ5 | frizzled family receptor 5 |
C00044491
|
| 8325 | FZD8, FZ-8, hFZ8 | frizzled family receptor 8 |
C00044491
|
| 55632 | G2E3, KIAA1333, PHF7B | G2/M-phase specific E3 ubiquitin protein ligase |
C00044491
|
| 10146 | G3BP1, G3BP, HDH-VIII | GTPase activating protein (SH3 domain) binding protein 1 (EC:3.6.4.13 3.6.4.12) |
C00044491
|
| 2539 | G6PD, G6PD1 | glucose-6-phosphate dehydrogenase (EC:1.1.1.49) |
C00044491
|
| 2548 | GAA, LYAG | glucosidase, alpha; acid (EC:3.2.1.20) |
C00044491
|
| 2549 | GAB1 | GRB2-associated binding protein 1 |
C00044491
|
| 11345 | GABARAPL2, ATG8, ATG8C, GATE-16, GATE16, GEF-2, GEF2 | GABA(A) receptor-associated protein-like 2 |
C00044491
|
| 2551 | GABPA, E4TF1-60, E4TF1A, NFT2, NRF2, NRF2A, RCH04A07 | GA binding protein transcription factor, alpha subunit 60kDa |
C00044491
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00044491
|
| 4616 | GADD45B, GADD45BETA, MYD118 | growth arrest and DNA-damage-inducible, beta |
C00044491
|
| 9514 | GAL3ST1, CST | galactose-3-O-sulfotransferase 1 (EC:2.8.2.11) |
C00044491
|
| 79690 | GAL3ST4, GAL3ST-4 | galactose-3-O-sulfotransferase 4 |
C00044491
|
| 2582 | GALE, SDR1E1 | UDP-galactose-4-epimerase (EC:5.1.3.2) |
C00044491
|
| 2585 | GALK2, GK2 | galactokinase 2 (EC:2.7.1.157) |
C00044491
|
| 130589 | GALM, IBD1 | galactose mutarotase (aldose 1-epimerase) (EC:5.1.3.3) |
C00044491
|
| 2588 | GALNS, GALNAC6S, GAS, GalN6S, MPS4A | galactosamine (N-acetyl)-6-sulfate sulfatase (EC:3.1.6.4) |
C00044491
|
| 63917 | GALNT11, GALNACT11 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) (EC:2.4.1.41) |
C00044491
|
| 2590 | GALNT2, GalNAc-T2 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) (EC:2.4.1.41) |
C00044491
|
| 50614 | GALNT9, GALNAC-T9, GALNACT9 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) (EC:2.4.1.41) |
C00044491
|
| 2587 | GALR1, GALNR, GALNR1 | galanin receptor 1 |
C00044491
|
| 54433 | GAR1, NOLA1 | GAR1 ribonucleoprotein |
C00044491
|
| 64762 | GAREM, C18orf11, FAM59A, Gm944 | GRB2 associated, regulator of MAPK1 |
C00044491
|
| 2618 | GART, AIRS, GARS, GARTF, PAIS, PGFT, PRGS | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase (EC:2.1.2.2 6.3.3.1 6.3.4.13) |
C00044491
|
| 2619 | GAS1 | growth arrest-specific 1 |
C00044491
|
| 2620 | GAS2 | growth arrest-specific 2 |
C00044491
|
| 283431 | GAS2L3, G2L3 | growth arrest-specific 2 like 3 |
C00044491
|
| 54815 | GATAD2A, p66alpha | GATA zinc finger domain containing 2A |
C00044491
|
| 57459 | GATAD2B, MRD18, P66beta, RP11-216N14.6, p68 | GATA zinc finger domain containing 2B |
C00044491
|
| 283459 | GATC, 15E1.2 | glutamyl-tRNA(Gln) amidotransferase, subunit C |
C00044491
|
| 2628 | GATM, AGAT, AT, CCDS3 | glycine amidinotransferase (L-arginine:glycine amidinotransferase) (EC:2.1.4.1) |
C00044491
|
| 2634 | GBP2 | guanylate binding protein 2, interferon-inducible |
C00044491
|
| 2635 | GBP3 | guanylate binding protein 3 |
C00044491
|
| 115361 | GBP4, Mpa2 | guanylate binding protein 4 |
C00044491
|
| 2637 | GBX2 | gastrulation brain homeobox 2 |
C00044491
|
| 2638 | GC, DBP, DBP/GC, GRD3, VDBG, VDBP | group-specific component (vitamin D binding protein) (EC:6.4.-.-) |
C00044491
|
| 9648 | GCC2, GCC185, RANBP2L4, REN53 | GRIP and coiled-coil domain containing 2 |
C00044491
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00044491
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00044491
|
| 9247 | GCM2, GCMB, hGCMb | glial cells missing homolog 2 (Drosophila) |
C00044491
|
| 10985 | GCN1L1, GCN1, GCN1L, PRIC295 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) |
C00044491
|
| 2651 | GCNT2, CCAT, CTRCT13, GCNT2C, GCNT5, IGNT, II, NACGT1, NAGCT1, ULG3, bA360O19.2, bA421M1.1 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) (EC:2.4.1.150) |
C00044491
|
| 9245 | GCNT3, C2/4GnT, C24GNT, C2GNT2, C2GNTM, GNTM | glucosaminyl (N-acetyl) transferase 3, mucin type (EC:2.4.1.102 2.4.1.150) |
C00044491
|
| 145781 | GCOM1, GRINL1A, Gcom2, MYZAP, MYZAP-POLR2M, gcom | GRINL1A complex locus 1 |
C00044491
|
| 2653 | GCSH, GCE, NKH | glycine cleavage system protein H (aminomethyl carrier) |
C00044491
|
| 9615 | GDA, CYPIN, GUANASE, NEDASIN | guanine deaminase (EC:3.5.4.3) |
C00044491
|
| 2664 | GDI1, 1A, GDIL, MRX41, MRX48, OPHN2, RABGD1A, RABGDIA, XAP-4 | GDP dissociation inhibitor 1 |
C00044491
|
| 284161 | GDPD1, GDE4 | glycerophosphodiester phosphodiesterase domain containing 1 |
C00044491
|
| 2669 | GEM, KIR | GTP binding protein overexpressed in skeletal muscle |
C00044491
|
| 79833 | GEMIN6 | gem (nuclear organelle) associated protein 6 |
C00044491
|
| 348654 | GEN1, Gen | GEN1 Holliday junction 5' flap endonuclease |
C00044491
|
| 85476 | GFM1, COXPD1, EFG, EFG1, EFGM, EGF1, GFM, hEFG1 | G elongation factor, mitochondrial 1 |
C00044491
|
| 23163 | GGA3 | golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
C00044491
|
| 8836 | GGH, GH | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (EC:3.4.19.9) |
C00044491
|
| 2686 | GGT7, D20S101, GGT4, GGTL3, GGTL5, dJ18C9.2 | gamma-glutamyltransferase 7 (EC:2.3.2.2 3.4.19.13) |
C00044491
|
| 2690 | GHR, GHBP | growth hormone receptor |
C00044491
|
| 26058 | GIGYF2, GYF2, PARK11, PERQ2, PERQ3, TNRC15 | GRB10 interacting GYF protein 2 |
C00044491
|
| 55303 | GIMAP4, IAN-1, IAN1, IMAP4 | GTPase, IMAP family member 4 |
C00044491
|
| 474344 | GIMAP6, IAN-2, IAN-6, IAN2, IAN6 | GTPase, IMAP family member 6 |
C00044491
|
| 116254 | GINM1, C6orf72, dJ12G14.2 | glycoprotein integral membrane 1 |
C00044491
|
| 9837 | GINS1, PSF1 | GINS complex subunit 1 (Psf1 homolog) |
C00044491
|
| 51659 | GINS2, HSPC037, PSF2, Pfs2 | GINS complex subunit 2 (Psf2 homolog) |
C00044491
|
| 64785 | GINS3, PSF3 | GINS complex subunit 3 (Psf3 homolog) |
C00044491
|
| 28964 | GIT1 | G protein-coupled receptor kinase interacting ArfGAP 1 |
C00044491
|
| 9815 | GIT2, CAT-2, CAT2 | G protein-coupled receptor kinase interacting ArfGAP 2 |
C00044491
|
| 2705 | GJB1, CMTX, CMTX1, CX32 | gap junction protein, beta 1, 32kDa |
C00044491
|
| 2706 | GJB2, CX26, DFNA3, DFNA3A, DFNB1, DFNB1A, HID, KID, NSRD1, PPK | gap junction protein, beta 2, 26kDa |
C00044491
|
| 219770 | GJD4, CX40.1, RP11-425A6.2 | gap junction protein, delta 4, 40.1kDa |
C00044491
|
| 2710 | GK, GK1, GKD | glycerol kinase (EC:2.7.1.30) |
C00044491
|
| 2713 | GK3P, GKP3, GKTB | glycerol kinase 3 pseudogene |
C00044491
|
| 2717 | GLA, GALA | galactosidase, alpha (EC:3.2.1.22) |
C00044491
|
| 79411 | GLB1L | galactosidase, beta 1-like |
C00044491
|
| 113263 | GLCCI1, FAM117C, GCTR, GIG18, TSSN1 | glucocorticoid induced transcript 1 |
C00044491
|
| 26035 | GLCE, HSEPI | glucuronic acid epimerase (EC:5.1.3.17) |
C00044491
|
| 2731 | GLDC, GCE, GCSP, HYGN1 | glycine dehydrogenase (decarboxylating) (EC:1.4.4.2) |
C00044491
|
| 2733 | GLE1, GLE1L, LCCS, LCCS1, hGLE1 | GLE1 RNA export mediator |
C00044491
|
| 2736 | GLI2, HPE9, THP1, THP2 | GLI family zinc finger 2 |
C00044491
|
| 11010 | GLIPR1, CRISP7, GLIPR, RTVP1 | GLI pathogenesis-related 1 |
C00044491
|
| 2745 | GLRX, GRX, GRX1 | glutaredoxin (thioltransferase) (EC:1.8.4.1) |
C00044491
|
| 2744 | GLS, AAD20, GAC, GAM, GLS1, KGA | glutaminase (EC:3.5.1.2) |
C00044491
|
| 27165 | GLS2, GA, GLS, LGA, hLGA | glutaminase 2 (liver, mitochondrial) (EC:3.5.1.2) |
C00044491
|
| 144423 | GLT1D1 | glycosyltransferase 1 domain containing 1 (EC:2.4.-.-) |
C00044491
|
| 55830 | GLT8D1, MSTP139 | glycosyltransferase 8 domain containing 1 |
C00044491
|
| 23506 | GLTSCR1L, KIAA0240 | GLTSCR1-like |
C00044491
|
| 2746 | GLUD1, GDH, GDH1, GLUD | glutamate dehydrogenase 1 (EC:1.4.1.3) |
C00044491
|
| 2752 | GLUL, GLNS, GS, PIG43, PIG59 | glutamate-ammonia ligase (EC:4.1.1.15 6.3.1.2) |
C00044491
|
| 84656 | GLYR1, BM045, HIBDL, N-PAC, NP60 | glyoxylate reductase 1 homolog (Arabidopsis) |
C00044491
|
| 2760 | GM2A, GM2-AP, SAP-3 | GM2 ganglioside activator |
C00044491
|
| 64395 | GMCL1, BTBD13, GCL, GCL1, SPATA29 | germ cell-less, spermatogenesis associated 1 |
C00044491
|
| 2762 | GMDS, GMD, SDR3E1 | GDP-mannose 4,6-dehydratase (EC:4.2.1.47) |
C00044491
|
| 10691 | GMEB1, P96PIF, PIF96 | glucocorticoid modulatory element binding protein 1 |
C00044491
|
| 2764 | GMFB, GMF | glia maturation factor, beta |
C00044491
|
| 9535 | GMFG, GMF-GAMMA | glia maturation factor, gamma |
C00044491
|
| 51053 | GMNN, Gem | geminin, DNA replication inhibitor |
C00044491
|
| 2768 | GNA12, NNX3, RMP, gep | guanine nucleotide binding protein (G protein) alpha 12 |
C00044491
|
| 10672 | GNA13, G13 | guanine nucleotide binding protein (G protein), alpha 13 |
C00044491
|
| 2770 | GNAI1, Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
C00044491
|
| 2771 | GNAI2, GIP, GNAI2B, H_LUCA15.1, H_LUCA16.1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
C00044491
|
| 10681 | GNB5, GB5 | guanine nucleotide binding protein (G protein), beta 5 |
C00044491
|
| 55970 | GNG12 | guanine nucleotide binding protein (G protein), gamma 12 |
C00044491
|
| 26354 | GNL3, C77032, E2IG3, NNP47, NS | guanine nucleotide binding protein-like 3 (nucleolar) |
C00044491
|
| 8443 | GNPAT, DAP-AT, DAPAT, DHAPAT | glyceronephosphate O-acyltransferase (EC:2.3.1.42) |
C00044491
|
| 64841 | GNPNAT1, GNPNAT, Gpnat1 | glucosamine-phosphate N-acetyltransferase 1 (EC:2.3.1.4) |
C00044491
|
| 84572 | GNPTG, C16orf27, GNPTAG, LP2537, RJD9 | N-acetylglucosamine-1-phosphate transferase, gamma subunit |
C00044491
|
| 2799 | GNS, G6S | glucosamine (N-acetyl)-6-sulfatase (EC:3.1.6.14) |
C00044491
|
| 2802 | GOLGA3, GCP170, MEA-2 | golgin A3 |
C00044491
|
| 64083 | GOLPH3, GOPP1, GPP34, MIDAS | golgi phosphoprotein 3 (coat-protein) |
C00044491
|
| 92344 | GORAB, GO, NTKLBP1, SCYL1BP1 | golgin, RAB6-interacting |
C00044491
|
| 2805 | GOT1, ASTQTL1, GIG18, cAspAT, cCAT | glutamic-oxaloacetic transaminase 1, soluble (EC:2.6.1.1 2.6.1.3) |
C00044491
|
| 2812 | GP1BB, BDPLT1, BS, CD42C, GPIBB | glycoprotein Ib (platelet), beta polypeptide |
C00044491
|
| 57678 | GPAM, GPAT, GPAT1 | glycerol-3-phosphate acyltransferase, mitochondrial (EC:2.3.1.15) |
C00044491
|
| 55094 | GPATCH1, ECGP, GPATC1 | G patch domain containing 1 |
C00044491
|
| 55668 | GPATCH2L, C14orf118 | G patch domain containing 2-like |
C00044491
|
| 54865 | GPATCH4, GPATC4 | G patch domain containing 4 |
C00044491
|
| 65056 | GPBP1, GPBP, SSH6, VASCULIN | GC-rich promoter binding protein 1 |
C00044491
|
| 2817 | GPC1, glypican | glypican 1 |
C00044491
|
| 2239 | GPC4, K-glypican | glypican 4 |
C00044491
|
| 10082 | GPC6, OMIMD1 | glypican 6 |
C00044491
|
| 56261 | GPCPD1, GDE5, GDPD6, PREI4 | glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) (EC:3.1.4.2) |
C00044491
|
| 2819 | GPD1, GPD-C, GPDH-C, HTGTI | glycerol-3-phosphate dehydrogenase 1 (soluble) (EC:1.1.1.8) |
C00044491
|
| 2852 | GPER1, CEPR, CMKRL2, DRY12, FEG-1, GPCR-Br, GPER, GPR30, LERGU, LERGU2, LyGPR | G protein-coupled estrogen receptor 1 |
C00044491
|
| 2821 | GPI, AMF, GNPI, NLK, PGI, PHI, SA-36, SA36 | glucose-6-phosphate isomerase (EC:5.3.1.9) |
C00044491
|
| 57211 | GPR126, APG1, DREG, PS1TP2, VIGR | G protein-coupled receptor 126 |
C00044491
|
| 283554 | GPR137C, TM7SF1L2 | G protein-coupled receptor 137C |
C00044491
|
| 124274 | GPR139, GPRg1, PGR3 | G protein-coupled receptor 139 |
C00044491
|
| 115330 | GPR146, PGR8 | G protein-coupled receptor 146 |
C00044491
|
| 80045 | GPR157 | G protein-coupled receptor 157 |
C00044491
|
| 27239 | GPR162, A-2, GRCA | G protein-coupled receptor 162 |
C00044491
|
| 2840 | GPR17 | G protein-coupled receptor 17 |
C00044491
|
| 11318 | GPR182, 7TMR, ADMR, AM-R, AMR, G10D, gamrh, hrhAMR | G protein-coupled receptor 182 |
C00044491
|
| 2843 | GPR20 | G protein-coupled receptor 20 |
C00044491
|
| 2828 | GPR4 | G protein-coupled receptor 4 |
C00044491
|
| 9289 | GPR56, BFPP, TM7LN4, TM7XN1 | G protein-coupled receptor 56 |
C00044491
|
| 222487 | GPR97, PB99, PGR26 | G protein-coupled receptor 97 |
C00044491
|
| 9052 | GPRC5A, GPCR5A, RAI3, RAIG1 | G protein-coupled receptor, family C, group 5, member A |
C00044491
|
| 51704 | GPRC5B, RAIG-2, RAIG2 | G protein-coupled receptor, family C, group 5, member B |
C00044491
|
| 29899 | GPSM2, CMCS, DFNB82, LGN, PINS | G-protein signaling modulator 2 |
C00044491
|
| 2877 | GPX2, GI-GPx, GPRP, GPRP-2, GPx-2, GPx-GI, GSHPX-GI, GSHPx-2 | glutathione peroxidase 2 (gastrointestinal) (EC:1.11.1.9) |
C00044491
|
| 2878 | GPX3, GPx-P, GSHPx-3, GSHPx-P | glutathione peroxidase 3 (plasma) (EC:1.11.1.9) |
C00044491
|
| 54762 | GRAMD1C | GRAM domain containing 1C |
C00044491
|
| 10750 | GRAP | GRB2-related adaptor protein |
C00044491
|
| 2888 | GRB14 | growth factor receptor-bound protein 14 |
C00044491
|
| 29841 | GRHL1, LBP32, MGR, NH32, TFCP2L2 | grainyhead-like 1 (Drosophila) |
C00044491
|
| 2907 | GRINA, HNRGW, LFG1, NMDARA1, TMBIM3 | glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
C00044491
|
| 2915 | GRM5, GPRC1E, MGLUR5, mGlu5 | glutamate receptor, metabotropic 5 |
C00044491
|
| 2896 | GRN, CLN11, GEP, GP88, PCDGF, PEPI, PGRN | granulin |
C00044491
|
| 79774 | GRTP1, TBC1D6 | growth hormone regulated TBC protein 1 |
C00044491
|
| 54103 | GSAP, PION | gamma-secretase activating protein |
C00044491
|
| 55876 | GSDMB, GSDML, PRO2521 | gasdermin B |
C00044491
|
| 23199 | GSE1, KIAA0182 | Gse1 coiled-coil protein |
C00044491
|
| 146395 | GSG1L, PRO19651 | GSG1-like |
C00044491
|
| 83903 | GSG2, HASPIN | germ cell associated 2 (haspin) (EC:2.7.11.1) |
C00044491
|
| 51527 | GSKIP, C14orf129 | GSK3B interacting protein |
C00044491
|
| 2935 | GSPT1, 551G9.2, ETF3A, GST1, eRF3a | G1 to S phase transition 1 |
C00044491
|
| 23708 | GSPT2, ERF3B, GST2 | G1 to S phase transition 2 |
C00044491
|
| 9446 | GSTO1, GSTO_1-1, GSTTLp28, P28, SPG-R | glutathione S-transferase omega 1 (EC:2.5.1.18 1.8.5.1 1.20.4.2) |
C00044491
|
| 2950 | GSTP1, DFN7, FAEES3, GST3, GSTP, PI | glutathione S-transferase pi 1 (EC:2.5.1.18) |
C00044491
|
| 219409 | GSX1, GSH1, Gsh-1 | GS homeobox 1 |
C00044491
|
| 2959 | GTF2B, TF2B, TFIIB | general transcription factor IIB |
C00044491
|
| 2965 | GTF2H1, BTF2, P62, TFB1, TFIIH | general transcription factor IIH, polypeptide 1, 62kDa |
C00044491
|
| 404672 | GTF2H5, C6orf175, TFB5, TFIIH, TGF2H5, TTD, TTD-A, TTDA, bA120J8.2 | general transcription factor IIH, polypeptide 5 |
C00044491
|
| 2969 | GTF2I, BAP135, BTKAP1, DIWS, GTFII-I, IB291, SPIN, TFII-I, WBS, WBSCR6 | general transcription factor IIi |
C00044491
|
| 2971 | GTF3A, AP2, TFIIIA | general transcription factor IIIA |
C00044491
|
| 2976 | GTF3C2, TFIIIC-BETA, TFIIIC110 | general transcription factor IIIC, polypeptide 2, beta 110kDa |
C00044491
|
| 9328 | GTF3C5, TFIIIC63, TFIIICepsilon, TFiiiC2-63 | general transcription factor IIIC, polypeptide 5, 63kDa |
C00044491
|
| 112495 | GTF3C6, C6orf51, TFIIIC35, bA397G5.3 | general transcription factor IIIC, polypeptide 6, alpha 35kDa |
C00044491
|
| 51512 | GTSE1, B99 | G-2 and S-phase expressed 1 |
C00044491
|
| 2978 | GUCA1A, C6orf131, COD3, CORD14, GCAP, GCAP1, GUCA, GUCA1, dJ139D8.6 | guanylate cyclase activator 1A (retina) |
C00044491
|
| 60558 | GUF1, EF-4 | GUF1 GTPase homolog (S. cerevisiae) |
C00044491
|
| 2990 | GUSB, BG, MPS7 | glucuronidase, beta (EC:3.2.1.31) |
C00044491
|
| 283464 | GXYLT1, GLT8D3 | glucoside xylosyltransferase 1 (EC:2.4.1.-) |
C00044491
|
| 27357 |
C00044491
|
||
| 2992 | GYG1, GSD15, GYG | glycogenin 1 (EC:2.4.1.186) |
C00044491
|
| 8908 | GYG2, GN-2, GN2 | glycogenin 2 (EC:2.4.1.186) |
C00044491
|
| 2995 | GYPC, CD236, CD236R, GE, GPC, GPD, GYPD, PAS-2, PAS-2' | glycophorin C (Gerbich blood group) |
C00044491
|
| 64412 | GZF1, ZBTB23, ZNF336 | GDNF-inducible zinc finger protein 1 |
C00044491
|
| 3001 | GZMA, CTLA3, HFSP | granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) (EC:3.4.21.78) |
C00044491
|
| 8971 | H1FX, H1X | H1 histone family, member X |
C00044491
|
| 55766 | H2AFJ, H2AJ | H2A histone family, member J |
C00044491
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00044491
|
| 9555 | H2AFY, H2A.y, H2A/y, H2AF12M, H2AFJ, MACROH2A1.1, mH2A1, macroH2A1.2 | H2A histone family, member Y |
C00044491
|
| 54145 |
C00044491
|
||
| 3021 | H3F3B, H3.3B | H3 histone, family 3B (H3.3B) |
C00044491
|
| 9563 | H6PD, CORTRD1, G6PDH, GDH | hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (EC:1.1.1.47 3.1.1.31) |
C00044491
|
| 3026 | HABP2, FSAP, HABP, HGFAL, PHBP | hyaluronan binding protein 2 |
C00044491
|
| 3032 | HADHB, ECHB, MTPB, TP-BETA | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit (EC:2.3.1.16) |
C00044491
|
| 84264 | HAGHL | hydroxyacylglutathione hydrolase-like (EC:3.1.2.6) |
C00044491
|
| 3034 | HAL, HIS, HSTD | histidine ammonia-lyase (EC:4.3.1.3) |
C00044491
|
| 57817 | HAMP, HEPC, HFE2B, LEAP1, PLTR | hepcidin antimicrobial peptide |
C00044491
|
| 60484 | HAPLN2, BRAL1 | hyaluronan and proteoglycan link protein 2 |
C00044491
|
| 8520 | HAT1, KAT1 | histone acetyltransferase 1 (EC:2.3.1.48) |
C00044491
|
| 115106 | HAUS1, CCDC5, HEI-C, HEIC, HsT1461 | HAUS augmin-like complex, subunit 1 |
C00044491
|
| 79441 | HAUS3, C4orf15, IT1, dgt3 | HAUS augmin-like complex, subunit 3 |
C00044491
|
| 54930 | HAUS4, C14orf94 | HAUS augmin-like complex, subunit 4 |
C00044491
|
| 1839 | HBEGF, DTR, DTS, DTSF, HEGFL | heparin-binding EGF-like growth factor |
C00044491
|
| 3042 | HBM, HBAP2 | hemoglobin, mu |
C00044491
|
| 26959 | HBP1 | HMG-box transcription factor 1 |
C00044491
|
| 3049 | HBQ1 | hemoglobin, theta 1 |
C00044491
|
| 8843 | HCAR3, GPR109B, HCA3, HM74, PUMAG, Puma-g | hydroxycarboxylic acid receptor 3 |
C00044491
|
| 29915 | HCFC2, HCF-2, HCF2 | host cell factor C2 |
C00044491
|
| 10866 | HCP5, 6S2650E, D6S2650E, P5-1 | HLA complex P5 (non-protein coding) |
C00044491
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00044491
|
| 51020 | HDDC2, C6orf74, NS5ATP2, dJ167O5.2 | HD domain containing 2 |
C00044491
|
| 374659 | HDDC3, (ppGpp)ase, MESH1 | HD domain containing 3 (EC:3.1.7.2) |
C00044491
|
| 3068 | HDGF, HMG1L2 | hepatoma-derived growth factor |
C00044491
|
| 55127 | HEATR1, BAP28, UTP10 | HEAT repeat containing 1 |
C00044491
|
| 54919 | HEATR2, CILD18 | HEAT repeat containing 2 |
C00044491
|
| 55027 | HEATR3, SYO1 | HEAT repeat containing 3 |
C00044491
|
| 25938 | HEATR5A, C14orf125 | HEAT repeat containing 5A |
C00044491
|
| 63897 | HEATR6, ABC1 | HEAT repeat containing 6 |
C00044491
|
| 3070 | HELLS, LSH, PASG, SMARCA6 | helicase, lymphoid-specific |
C00044491
|
| 9931 | HELZ, DHRC, DRHC, HUMORF5 | helicase with zinc finger |
C00044491
|
| 85441 | HELZ2, PDIP-1, PRIC285 | helicase with zinc finger 2, transcriptional coactivator |
C00044491
|
| 8916 | HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
C00044491
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00044491
|
| 57801 | HES4, bHLHb42 | hes family bHLH transcription factor 4 |
C00044491
|
| 388585 | HES5, bHLHb38 | hes family bHLH transcription factor 5 |
C00044491
|
| 3074 | HEXB, ENC-1AS | hexosaminidase B (beta polypeptide) (EC:3.2.1.52) |
C00044491
|
| 10614 | HEXIM1, CLP1, EDG1, HIS1, MAQ1 | hexamethylene bis-acetamide inducible 1 |
C00044491
|
| 23493 | HEY2, CHF1, GRIDLOCK, GRL, HERP1, HESR2, HRT2, bHLHb32 | hes-related family bHLH transcription factor with YRPW motif 2 |
C00044491
|
| 100128124 | HGC6.3 | uncharacterized LOC100128124 |
C00044491
|
| 138050 | HGSNAT, HGNAT, MPS3C, TMEM76 | heparan-alpha-glucosaminide N-acetyltransferase (EC:2.3.1.78) |
C00044491
|
| 84439 | HHIPL1, KIAA1822, UNQ9245 | HHIP-like 1 |
C00044491
|
| 11147 | HHLA3 | HERV-H LTR-associating 3 |
C00044491
|
| 23119 | HIC2, HRG22, ZBTB30, ZNF907 | hypermethylated in cancer 2 |
C00044491
|
| 55662 | HIF1AN, FIH1 | hypoxia inducible factor 1, alpha subunit inhibitor (EC:1.14.11.30 1.14.11.n4) |
C00044491
|
| 3094 | HINT1, HINT, NMAN, PKCI-1, PRKCNH1 | histidine triad nucleotide binding protein 1 |
C00044491
|
| 3092 | HIP1, HIP-I, ILWEQ | huntingtin interacting protein 1 |
C00044491
|
| 9026 | HIP1R, HIP12, HIP3, ILWEQ | huntingtin interacting protein 1 related |
C00044491
|
| 28996 | HIPK2, PRO0593 | homeodomain interacting protein kinase 2 (EC:2.7.11.1) |
C00044491
|
| 3006 | HIST1H1C, H1.2, H1C, H1F2, H1s-1 | histone cluster 1, H1c |
C00044491
|
| 3010 | HIST1H1T, H1FT, H1t, dJ221C16.2 | histone cluster 1, H1t |
C00044491
|
| 8334 | HIST1H2AC, H2A/l, H2AFL, dJ221C16.4 | histone cluster 1, H2ac |
C00044491
|
| 3018 | HIST1H2BB, H2B.1, H2B/f, H2BFF | histone cluster 1, H2bb |
C00044491
|
| 8347 | HIST1H2BC, H2B.1, H2B/l, H2BFL, dJ221C16.3 | histone cluster 1, H2bc |
C00044491
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00044491
|
| 8345 | HIST1H2BH, H2B/j, H2BFJ | histone cluster 1, H2bh |
C00044491
|
| 8970 | HIST1H2BJ, H2B/r, H2BFR, H2BJ | histone cluster 1, H2bj |
C00044491
|
| 85236 | HIST1H2BK, H2B/S, H2BFAiii, H2BFT, H2BK | histone cluster 1, H2bk |
C00044491
|
| 8340 | HIST1H2BL, H2B/c, H2BFC, dJ97D16.4 | histone cluster 1, H2bl |
C00044491
|
| 8342 | HIST1H2BM, H2B/e, H2BFE, dJ160A22.3 | histone cluster 1, H2bm |
C00044491
|
| 8363 | HIST1H4J, H4/e, H4F2iv, H4FE, dJ160A22.2 | histone cluster 1, H4j |
C00044491
|
| 8337 | HIST2H2AA3, H2A, H2A.2, H2A/O, H2A/q, H2AFO, H2a-615, HIST2H2AA | histone cluster 2, H2aa3 |
C00044491
|
| 8349 | HIST2H2BE, GL105, H2B, H2B.1, H2BFQ, H2BGL105, H2BQ | histone cluster 2, H2be |
C00044491
|
| 55355 | HJURP, FAKTS, URLC9, hFLEG1 | Holliday junction recognition protein |
C00044491
|
| 3134 | HLA-F, CDA12, HLA-5.4, HLA-CDA12, HLAF | major histocompatibility complex, class I, F |
C00044491
|
| 6596 | HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3, ZBU1 | helicase-like transcription factor |
C00044491
|
| 10363 | HMG20A, HMGX1, HMGXB1 | high mobility group 20A |
C00044491
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00044491
|
| 3155 | HMGCL, HL | 3-hydroxymethyl-3-methylglutaryl-CoA lyase (EC:4.1.3.4) |
C00044491
|
| 3157 | HMGCS1, HMGCS | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) (EC:2.3.3.10) |
C00044491
|
| 3151 | HMGN2, HMG17 | high mobility group nucleosomal binding domain 2 |
C00044491
|
| 3161 | HMMR, CD168, IHABP, RHAMM | hyaluronan-mediated motility receptor (RHAMM) |
C00044491
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00044491
|
| 51155 | HN1, ARM2, HN1A | hematological and neurological expressed 1 |
C00044491
|
| 6928 | HNF1B, FJHN, HNF-1B, HNF1beta, HNF2, HPC11, LF-B3, LFB3, MODY5, TCF-2, TCF2, VHNF1 | HNF1 homeobox B |
C00044491
|
| 3174 | HNF4G, NR2A2, NR2A3 | hepatocyte nuclear factor 4, gamma |
C00044491
|
| 3176 | HNMT, HMT, HNMT-S1, HNMT-S2 | histamine N-methyltransferase (EC:2.1.1.8) |
C00044491
|
| 3178 | HNRNPA1, ALS19, ALS20, HNRPA1, HNRPA1L3, IBMPFD3, hnRNP_A1, hnRNP-A1 | heterogeneous nuclear ribonucleoprotein A1 |
C00044491
|
| 3181 | HNRNPA2B1, HNRNPA2, HNRNPB1, HNRPA2, HNRPA2B1, HNRPB1, IBMPFD2, RNPA2, SNRPB1 | heterogeneous nuclear ribonucleoprotein A2/B1 |
C00044491
|
| 3182 | HNRNPAB, ABBP1, HNRPAB | heterogeneous nuclear ribonucleoprotein A/B |
C00044491
|
| 3184 | HNRNPD, AUF1, AUF1A, HNRPD, P37, hnRNPD0 | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
C00044491
|
| 3189 | HNRNPH3, 2H9, HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) |
C00044491
|
| 4670 | HNRNPM, CEAR, HNRNPM4, HNRPM, HNRPM4, HTGR1, NAGR1, hnRNP_M | heterogeneous nuclear ribonucleoprotein M |
C00044491
|
| 11100 | HNRNPUL1, E1B-AP5, E1BAP5, HNRPUL1 | heterogeneous nuclear ribonucleoprotein U-like 1 |
C00044491
|
| 379948 |
C00044491
|
||
| 9456 | HOMER1, HOMER, HOMER1A, HOMER1B, HOMER1C, SYN47, Ves-1 | homer homolog 1 (Drosophila) |
C00044491
|
| 51361 | HOOK1, HK1 | hook microtubule-tethering protein 1 |
C00044491
|
| 3238 | HOXD12, HOX4H | homeobox D12 |
C00044491
|
| 50809 | HP1BP3, HP1-BP74 | heterochromatin protein 1, binding protein 3 |
C00044491
|
| 3242 | HPD, 4-HPPD, 4HPPD, GLOD3, HPPDASE, PPD | 4-hydroxyphenylpyruvate dioxygenase (EC:1.13.11.27) |
C00044491
|
| 3249 | HPN, TMPRSS1 | hepsin (EC:3.4.21.106) |
C00044491
|
| 11234 | HPS5, AIBP63 | Hermansky-Pudlak syndrome 5 |
C00044491
|
| 10855 | HPSE, HPA, HPA1, HPR1, HPSE1, HSE1 | heparanase (EC:3.2.1.166) |
C00044491
|
| 55806 | HR, ALUNC, AU, HSA277165, MUHH, MUHH1 | hair growth associated |
C00044491
|
| 9953 | HS3ST3B1, 30ST3B1, 3OST3B1 | heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 (EC:2.8.2.30) |
C00044491
|
| 51170 | HSD17B11, 17BHSD11, DHRS8, PAN1B, RETSDR2, SDR16C2 | hydroxysteroid (17-beta) dehydrogenase 11 (EC:1.1.1.62) |
C00044491
|
| 3294 | HSD17B2, EDH17B2, HSD17, SDR9C2 | hydroxysteroid (17-beta) dehydrogenase 2 (EC:1.1.1.62 1.1.1.239) |
C00044491
|
| 8630 | HSD17B6, HSE, RODH, SDR9C6 | hydroxysteroid (17-beta) dehydrogenase 6 (EC:1.1.1.62 1.1.1.239 1.1.1.105) |
C00044491
|
| 80270 | HSD3B7, CBAS1, PFIC4, SDR11E3 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 (EC:1.1.1.181) |
C00044491
|
| 3297 | HSF1, HSTF1 | heat shock transcription factor 1 |
C00044491
|
| 3320 | HSP90AA1, EL52, HSP86, HSP89A, HSP90A, HSP90N, HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 |
C00044491
|
| 3324 |
C00044491
|
||
| 3326 | HSP90AB1, D6S182, HSP84, HSP90B, HSPC2, HSPCB | heat shock protein 90kDa alpha (cytosolic), class B member 1 |
C00044491
|
| 391634 |
C00044491
|
||
| 7184 | HSP90B1, ECGP, GP96, GRP94, TRA1 | heat shock protein 90kDa beta (Grp94), member 1 |
C00044491
|
| 116835 | HSPA12B, C20orf60, dJ1009E24.2 | heat shock 70kD protein 12B |
C00044491
|
| 6782 | HSPA13, STCH | heat shock protein 70kDa family, member 13 |
C00044491
|
| 3305 | HSPA1L, HSP70-1L, HSP70-HOM, HSP70T, hum70t | heat shock 70kDa protein 1-like |
C00044491
|
| 3306 | HSPA2, HSP70-2, HSP70-3 | heat shock 70kDa protein 2 |
C00044491
|
| 22824 | HSPA4L, APG-1, HSPH3, Osp94 | heat shock 70kDa protein 4-like |
C00044491
|
| 3310 | HSPA6 | heat shock 70kDa protein 6 (HSP70B') |
C00044491
|
| 3312 | HSPA8, HSC54, HSC70, HSC71, HSP71, HSP73, HSPA10, LAP1, NIP71 | heat shock 70kDa protein 8 |
C00044491
|
| 79663 | HSPBAP1, PASS1 | HSPB (heat shock 27kDa) associated protein 1 |
C00044491
|
| 23640 | HSPBP1, FES1 | HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
C00044491
|
| 3329 | HSPD1, CPN60, GROEL, HLD4, HSP-60, HSP60, HSP65, HuCHA60, SPG13 | heat shock 60kDa protein 1 (chaperonin) |
C00044491
|
| 10808 | HSPH1, HSP105, HSP105A, HSP105B, NY-CO-25 | heat shock 105kDa/110kDa protein 1 |
C00044491
|
| 3351 | HTR1B, 5-HT1B, 5-HT1DB, HTR1D2, HTR1DB, S12 | 5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
C00044491
|
| 5654 | HTRA1, ARMD7, CARASIL, HtrA, L56, ORF480, PRSS11 | HtrA serine peptidase 1 (EC:3.4.21.-) |
C00044491
|
| 219844 | HYLS1, HLS | hydrolethalus syndrome 1 |
C00044491
|
| 55699 | IARS2, ILERS | isoleucyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.5) |
C00044491
|
| 25998 | IBTK, BTBD26, BTKI, RP1-93K22.1 | inhibitor of Bruton agammaglobulinemia tyrosine kinase |
C00044491
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00044491
|
| 3384 | ICAM2, CD102 | intercellular adhesion molecule 2 |
C00044491
|
| 3386 | ICAM4, CD242, LW | intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
C00044491
|
| 22858 | ICK, ECO, LCK2, MRK | intestinal cell (MAK-like) kinase (EC:2.7.11.22) |
C00044491
|
| 3397 | ID1, ID, bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
C00044491
|
| 3398 | ID2, GIG8, ID2A, ID2H, bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
C00044491
|
| 3417 | IDH1, IDCD, IDH, IDP, IDPC, PICD | isocitrate dehydrogenase 1 (NADP+), soluble (EC:1.1.1.42) |
C00044491
|
| 3422 | IDI1, IPP1, IPPI1 | isopentenyl-diphosphate delta isomerase 1 (EC:5.3.3.2) |
C00044491
|
| 414328 | IDNK, C9orf103, bA522I20.2 | idnK, gluconokinase homolog (E. coli) (EC:2.7.1.12) |
C00044491
|
| 9592 | IER2, ETR101 | immediate early response 2 |
C00044491
|
| 51278 | IER5, SBBI48 | immediate early response 5 |
C00044491
|
| 3428 | IFI16, IFNGIP1, PYHIN2 | interferon, gamma-inducible protein 16 |
C00044491
|
| 122509 | IFI27L1, FAM14B, ISG12C | interferon, alpha-inducible protein 27-like 1 |
C00044491
|
| 10561 | IFI44, MTAP44, TLDC5, p44 | interferon-induced protein 44 |
C00044491
|
| 2537 | IFI6, 6-16, FAM14C, G1P3, IFI-6-16, IFI616 | interferon, alpha-inducible protein 6 |
C00044491
|
| 64135 | IFIH1, Hlcd, IDDM19, MDA-5, MDA5, RLR-2 | interferon induced with helicase C domain 1 (EC:3.6.4.13) |
C00044491
|
| 3434 | IFIT1, C56, G10P1, IFI-56, IFI-56K, IFI56, IFIT-1, IFNAI1, ISG56, P56, RNM561 | interferon-induced protein with tetratricopeptide repeats 1 |
C00044491
|
| 3433 | IFIT2, G10P2, GARG-39, IFI-54, IFI-54K, IFI54, IFIT-2, ISG-54_K, ISG-54K, ISG54, P54, cig42 | interferon-induced protein with tetratricopeptide repeats 2 |
C00044491
|
| 3437 | IFIT3, CIG-49, GARG-49, IFI60, IFIT4, IRG2, ISG60, P60, RIG-G | interferon-induced protein with tetratricopeptide repeats 3 |
C00044491
|
| 8519 | IFITM1, 9-27, CD225, DSPA2a, IFI17, LEU13 | interferon induced transmembrane protein 1 |
C00044491
|
| 10581 | IFITM2, 1-8D, DSPA2c | interferon induced transmembrane protein 2 |
C00044491
|
| 10410 | IFITM3, 1-8U, DSPA2b, IP15 | interferon induced transmembrane protein 3 |
C00044491
|
| 3454 | IFNAR1, AVP, IFN-alpha-REC, IFNAR, IFNBR, IFRC | interferon (alpha, beta and omega) receptor 1 |
C00044491
|
| 3459 | IFNGR1, CD119, IFNGR | interferon gamma receptor 1 |
C00044491
|
| 3460 | IFNGR2, AF-1, IFGR2, IFNGT1 | interferon gamma receptor 2 (interferon gamma transducer 1) |
C00044491
|
| 3475 | IFRD1, PC4, TIS7 | interferon-related developmental regulator 1 |
C00044491
|
| 80173 | IFT74, CCDC2, CMG-1, CMG1 | intraflagellar transport 74 homolog (Chlamydomonas) |
C00044491
|
| 57560 | IFT80, ATD2, WDR56 | intraflagellar transport 80 homolog (Chlamydomonas) |
C00044491
|
| 8100 | IFT88, D13S1056E, DAF19, TG737, TTC10, hTg737 | intraflagellar transport 88 homolog (Chlamydomonas) |
C00044491
|
| 3482 | IGF2R, CD222, CIMPR, M6P-R, MPR1, MPRI | insulin-like growth factor 2 receptor |
C00044491
|
| 3484 | IGFBP1, AFBP, IBP1, IGF-BP25, PP12, hIGFBP-1 | insulin-like growth factor binding protein 1 |
C00044491
|
| 283284 | IGSF22 | immunoglobulin superfamily, member 22 |
C00044491
|
| 10261 | IGSF6, DORA | immunoglobulin superfamily, member 6 |
C00044491
|
| 121457 | IKBIP, IKIP | IKBKB interacting protein |
C00044491
|
| 8518 | IKBKAP, DYS, ELP1, FD, IKAP, IKI3, TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
C00044491
|
| 3551 | IKBKB, IKK-beta, IKK2, IKKB, NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (EC:2.7.11.10) |
C00044491
|
| 8517 | IKBKG, AMCBX1, FIP-3, FIP3, Fip3p, IKK-gamma, IP, IP1, IP2, IPD2, NEMO, ZC2HC9 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
C00044491
|
| 64376 | IKZF5, PEGASUS, ZNFN1A5 | IKAROS family zinc finger 5 (Pegasus) |
C00044491
|
| 3588 | IL10RB, CDW210B, CRF2-4, CRFB4, D21S58, D21S66, IL-10R2 | interleukin 10 receptor, beta |
C00044491
|
| 3590 | IL11RA, CRSDA | interleukin 11 receptor, alpha |
C00044491
|
| 3601 | IL15RA, CD215 | interleukin 15 receptor, alpha |
C00044491
|
| 27189 | IL17C, CX2, IL-17C, IL-21 | interleukin 17C |
C00044491
|
| 53342 | IL17D, IL-17D, IL-22, IL-27, IL27 | interleukin 17D |
C00044491
|
| 55540 | IL17RB, CRL4, EVI27, IL17BR, IL17RH1 | interleukin 17 receptor B |
C00044491
|
| 132014 | IL17RE | interleukin 17 receptor E |
C00044491
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00044491
|
| 3554 | IL1R1, CD121A, D2S1473, IL-1R-alpha, IL1R, IL1RA, P80 | interleukin 1 receptor, type I |
C00044491
|
| 27178 | IL37, FIL1, FIL1(ZETA), FIL1Z, IL-1F7, IL-1H, IL-1H4, IL-1RP1, IL-37, IL1F7, IL1H4, IL1RP1 | interleukin 37 |
C00044491
|
| 3570 | IL6R, CD126, IL-6R-1, IL-6RA, IL6Q, IL6RA, IL6RQ, gp80 | interleukin 6 receptor |
C00044491
|
| 3572 | IL6ST, CD130, CDW130, GP130, IL-6RB | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
C00044491
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00044491
|
| 3611 | ILK, ILK-1, ILK-2, P59, p59ILK | integrin-linked kinase (EC:2.7.11.1) |
C00044491
|
| 55364 | IMPACT, RWDD5 | impact RWD domain protein |
C00044491
|
| 54928 | IMPAD1, GPAPP, IMP_3, IMP-3, IMPA3 | inositol monophosphatase domain containing 1 (EC:3.1.3.7 3.1.3.25) |
C00044491
|
| 3614 | IMPDH1, IMPD, IMPD1, LCA11, RP10, sWSS2608 | IMP (inosine 5'-monophosphate) dehydrogenase 1 (EC:1.1.1.205) |
C00044491
|
| 10207 | INADL, Cipp, PATJ, hINADL | InaD-like (Drosophila) |
C00044491
|
| 3621 | ING1, p24ING1c, p33, p33ING1, p33ING1b, p47, p47ING1a | inhibitor of growth family, member 1 |
C00044491
|
| 3622 | ING2, ING1L, p33ING2 | inhibitor of growth family, member 2 |
C00044491
|
| 3623 | INHA | inhibin, alpha |
C00044491
|
| 3624 | INHBA, EDF, FRP | inhibin, beta A |
C00044491
|
| 3625 | INHBB | inhibin, beta B |
C00044491
|
| 83729 | INHBE | inhibin, beta E |
C00044491
|
| 54891 | INO80D | INO80 complex subunit D |
C00044491
|
| 3628 | INPP1 | inositol polyphosphate-1-phosphatase (EC:3.1.3.57) |
C00044491
|
| 3631 | INPP4A, INPP4, TVAS1 | inositol polyphosphate-4-phosphatase, type I, 107kDa (EC:3.1.3.66) |
C00044491
|
| 3632 | INPP5A, 5PTASE | inositol polyphosphate-5-phosphatase, 40kDa (EC:3.1.3.56) |
C00044491
|
| 3635 | INPP5D, SHIP, SHIP-1, SHIP1, SIP-145, hp51CN | inositol polyphosphate-5-phosphatase, 145kDa (EC:3.1.3.86) |
C00044491
|
| 51141 | INSIG2 | insulin induced gene 2 |
C00044491
|
| 3640 | INSL3, RLF, RLNL, ley-I-L | insulin-like 3 (Leydig cell) |
C00044491
|
| 3642 | INSM1, IA-1, IA1 | insulinoma-associated 1 |
C00044491
|
| 3643 | INSR, CD220, HHF5 | insulin receptor (EC:2.7.10.1) |
C00044491
|
| 55174 | INTS10, C8orf35, INT10 | integrator complex subunit 10 |
C00044491
|
| 65123 | INTS3, C1orf193, C1orf60, INT3, SOSS-A | integrator complex subunit 3 |
C00044491
|
| 26512 | INTS6, DBI-1, DDX26, DDX26A, DICE1, HDB, INT6, Notchl2 | integrator complex subunit 6 |
C00044491
|
| 25896 | INTS7, C1orf73, INT7 | integrator complex subunit 7 |
C00044491
|
| 117283 | IP6K3, IHPK3, INSP6K3 | inositol hexakisphosphate kinase 3 (EC:2.7.4.21) |
C00044491
|
| 51194 | IPO11, RanBP11 | importin 11 |
C00044491
|
| 9670 | IPO13, IMP13, KAP13, LGL2, RANBP13 | importin 13 |
C00044491
|
| 79711 | IPO4, Imp4 | importin 4 |
C00044491
|
| 3843 | IPO5, IMB3, KPNB3, Pse1, RANBP5, imp5 | importin 5 |
C00044491
|
| 10527 | IPO7, Imp7, RANBP7 | importin 7 |
C00044491
|
| 10526 | IPO8, RANBP8 | importin 8 |
C00044491
|
| 55705 | IPO9, Imp9 | importin 9 |
C00044491
|
| 124152 | IQCK | IQ motif containing K |
C00044491
|
| 10788 | IQGAP2 | IQ motif containing GTPase activating protein 2 |
C00044491
|
| 128239 | IQGAP3 | IQ motif containing GTPase activating protein 3 |
C00044491
|
| 9922 | IQSEC1, ARF-GEP100, ARFGEP100, BRAG2, GEP100 | IQ motif and Sec7 domain 1 |
C00044491
|
| 440073 | IQSEC3 | IQ motif and Sec7 domain 3 |
C00044491
|
| 3654 | IRAK1, IRAK, pelle | interleukin-1 receptor-associated kinase 1 (EC:2.7.11.1) |
C00044491
|
| 11213 | IRAK3, ASRT5, IRAKM | interleukin-1 receptor-associated kinase 3 (EC:2.7.11.1) |
C00044491
|
| 3658 | IREB2, ACO3, IRP2, IRP2AD | iron-responsive element binding protein 2 |
C00044491
|
| 3660 | IRF2, IRF-2 | interferon regulatory factor 2 |
C00044491
|
| 359948 | IRF2BP2 | interferon regulatory factor 2 binding protein 2 |
C00044491
|
| 64207 | IRF2BPL, C14orf4, EAP1 | interferon regulatory factor 2 binding protein-like |
C00044491
|
| 3661 | IRF3 | interferon regulatory factor 3 |
C00044491
|
| 3662 | IRF4, LSIRF, MUM1, NF-EM5 | interferon regulatory factor 4 |
C00044491
|
| 3665 | IRF7, IRF-7H, IRF7A, IRF7B, IRF7C, IRF7H | interferon regulatory factor 7 |
C00044491
|
| 10379 | IRF9, IRF-9, ISGF3, ISGF3G, p48 | interferon regulatory factor 9 |
C00044491
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00044491
|
| 8660 | IRS2, IRS-2 | insulin receptor substrate 2 |
C00044491
|
| 81689 | ISCA1, HBLD2, ISA1, RP11-507D14.2, hIscA | iron-sulfur cluster assembly 1 (EC:2.7.7.52) |
C00044491
|
| 23479 | ISCU, 2310020H20Rik, HML, ISU2, NIFU, NIFUN, hnifU | iron-sulfur cluster assembly enzyme |
C00044491
|
| 3669 | ISG20, CD25, HEM45 | interferon stimulated exonuclease gene 20kDa (EC:3.1.13.1) |
C00044491
|
| 51477 | ISYNA1, INO1, INOS, IPS, IPS_1, IPS-1 | inositol-3-phosphate synthase 1 (EC:5.5.1.4) |
C00044491
|
| 83737 | ITCH, AIF4, AIP4, NAPP1, dJ468O1.1 | itchy E3 ubiquitin protein ligase |
C00044491
|
| 81533 | ITFG1, TIP | integrin alpha FG-GAP repeat containing 1 |
C00044491
|
| 83986 | ITFG3, C16orf9, gs19 | integrin alpha FG-GAP repeat containing 3 |
C00044491
|
| 3675 | ITGA3, CD49C, GAP-B3, GAPB3, ILNEB, MSK18, VCA-2, VL3A, VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
C00044491
|
| 3685 | ITGAV, CD51, MSK8, VNRA, VTNR | integrin, alpha V |
C00044491
|
| 3693 | ITGB5 | integrin, beta 5 |
C00044491
|
| 81618 | ITM2C, BRI3, BRICD2C, E25, E25C, ITM3 | integral membrane protein 2C |
C00044491
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00044491
|
| 3709 | ITPR2, IP3R2 | inositol 1,4,5-trisphosphate receptor, type 2 |
C00044491
|
| 162073 | ITPRIPL2 | inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
C00044491
|
| 50618 | ITSN2, PRO2015, SH3D1B, SH3P18, SWA, SWAP | intersectin 2 |
C00044491
|
| 284359 | IZUMO1, IZUMO | izumo sperm-egg fusion 1 |
C00044491
|
| 9767 | JADE3, JADE-3, PHF16 | jade family PHD finger 3 |
C00044491
|
| 3716 | JAK1, JAK1A, JAK1B, JTK3 | Janus kinase 1 (EC:2.7.10.2) |
C00044491
|
| 3718 | JAK3, JAK-3, JAK3_HUMAN, JAKL, L-JAK, LJAK | Janus kinase 3 (EC:2.7.10.2) |
C00044491
|
| 80853 | KDM7A, JHDM1D | lysine (K)-specific demethylase 7A |
C00044491
|
| 221037 | JMJD1C, RP11-10C13.2, TRIP8 | jumonji domain containing 1C |
C00044491
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00044491
|
| 3726 | JUNB, AP-1 | jun B proto-oncogene |
C00044491
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00044491
|
| 23189 | KANK1, ANKRD15, CPSQ2, KANK | KN motif and ankyrin repeat domains 1 |
C00044491
|
| 25959 | KANK2, ANKRD25, MXRA3, SIP | KN motif and ankyrin repeat domains 2 |
C00044491
|
| 284058 | KANSL1, CENP-36, KDVS, KIAA1267, MSL1v1, NSL1, hMSL1v1 | KAT8 regulatory NSL complex subunit 1 |
C00044491
|
| 54934 | KANSL2, C12orf41, NSL2 | KAT8 regulatory NSL complex subunit 2 |
C00044491
|
| 3735 | KARS, CMTRIB, DFNB89, KARS1, KARS2, KRS | lysyl-tRNA synthetase (EC:6.1.1.6) |
C00044491
|
| 8850 | KAT2B, CAF, P, P/CAF, PCAF | K(lysine) acetyltransferase 2B (EC:2.3.1.48) |
C00044491
|
| 11104 | KATNA1 | katanin p60 (ATPase containing) subunit A 1 (EC:3.6.4.3) |
C00044491
|
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 (EC:3.6.4.3) |
C00044491
|
| 79768 | KATNBL1, C15orf29 | katanin p80 subunit B-like 1 |
C00044491
|
| 84541 | KBTBD8, TA-KRP | kelch repeat and BTB (POZ) domain containing 8 |
C00044491
|
| 3747 | KCNC2, KV3.2 | potassium voltage-gated channel, Shaw-related subfamily, member 2 |
C00044491
|
| 3752 | KCND3, KCND3L, KCND3S, KSHIVB, KV4.3, SCA19, SCA22 | potassium voltage-gated channel, Shal-related subfamily, member 3 |
C00044491
|
| 9992 | KCNE2, ATFB4, LQT5, LQT6, MIRP1 | potassium voltage-gated channel, Isk-related family, member 2 |
C00044491
|
| 3767 | KCNJ11, BIR, HHF2, IKATP, KIR6.2, PHHI, TNDM3 | potassium inwardly-rectifying channel, subfamily J, member 11 |
C00044491
|
| 3761 | KCNJ4, HIR, HIRK2, HRK1, IRK-3, IRK3, Kir2.3 | potassium inwardly-rectifying channel, subfamily J, member 4 |
C00044491
|
| 3775 | KCNK1, DPK, HOHO, K2P1, K2p1.1, KCNO1, TWIK-1, TWIK1 | potassium channel, subfamily K, member 1 |
C00044491
|
| 8645 | KCNK5, K2p5.1, TASK-2, TASK2 | potassium channel, subfamily K, member 5 |
C00044491
|
| 3782 | KCNN3, KCa2.3, SK3, SKCA3, hSK3 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
C00044491
|
| 3783 | KCNN4, IK1, IKCA1, KCA4, KCa3.1, SK4, hIKCa1, hKCa4, hSK4 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
C00044491
|
| 10984 | KCNQ1OT1, KCNQ1-AS2, KCNQ10T1, KvDMR1, KvLQT1-AS, LIT1, NCRNA00012 | KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
C00044491
|
| 3785 | KCNQ2, BFNC, BFNS1, EBN, EBN1, EIEE7, ENB1, HNSPC, KCNA11, KV7.2, KVEBN1 | potassium voltage-gated channel, KQT-like subfamily, member 2 |
C00044491
|
| 57582 | KCNT1, EIEE14, ENFL5, KCa4.1, SLACK, bA100C15.2 | potassium channel, subfamily T, member 1 |
C00044491
|
| 343450 | KCNT2, KCa4.2, SLICK, SLO2.1 | potassium channel, subfamily T, member 2 |
C00044491
|
| 83892 | KCTD10, BTBD28, ULRO61, hBACURD3 | potassium channel tetramerization domain containing 10 |
C00044491
|
| 147040 | KCTD11, C17orf36, KCASH1, REN, REN/KCTD11 | potassium channel tetramerization domain containing 11 |
C00044491
|
| 253980 | KCTD13, PDIP1, POLDIP1, hBACURD1 | potassium channel tetramerization domain containing 13 |
C00044491
|
| 65987 | KCTD14 | potassium channel tetramerization domain containing 14 |
C00044491
|
| 222658 | KCTD20, C6orf69, dJ108K11.3 | potassium channel tetramerization domain containing 20 |
C00044491
|
| 79070 | KDELC1, EP58, KDEL1 | KDEL (Lys-Asp-Glu-Leu) containing 1 |
C00044491
|
| 143888 | KDELC2 | KDEL (Lys-Asp-Glu-Leu) containing 2 |
C00044491
|
| 221656 | KDM1B, AOF1, C6orf193, LSD2, bA204B7.3, dJ298J15.2 | lysine (K)-specific demethylase 1B |
C00044491
|
| 22992 | KDM2A, CXXC8, FBL11, FBL7, FBXL11, JHDM1A, LILINA | lysine (K)-specific demethylase 2A (EC:1.14.11.27) |
C00044491
|
| 84678 | KDM2B, CXXC2, FBXL10, Fbl10, JHDM1B, PCCX2 | lysine (K)-specific demethylase 2B (EC:1.14.11.27) |
C00044491
|
| 51780 | KDM3B, 5qNCA, C5orf7, JMJD1B, NET22 | lysine (K)-specific demethylase 3B |
C00044491
|
| 23081 | KDM4C, GASC1, JHDM3C, JMJD2C, TDRD14C, bA146B14.1 | lysine (K)-specific demethylase 4C (EC:1.14.11.-) |
C00044491
|
| 2531 | KDSR, DHSR, FVT1, SDR35C1 | 3-ketodihydrosphingosine reductase (EC:1.1.1.102) |
C00044491
|
| 9817 | KEAP1, INrf2, KLHL19 | kelch-like ECH-associated protein 1 |
C00044491
|
| 10657 | KHDRBS1, Sam68, p62, p68 | KH domain containing, RNA binding, signal transduction associated 1 |
C00044491
|
| 10656 | KHDRBS3, Etle, SALP, SLM-2, SLM2, T-STAR, TSTAR, etoile | KH domain containing, RNA binding, signal transduction associated 3 |
C00044491
|
| 3795 | KHK | ketohexokinase (fructokinase) (EC:2.7.1.3) |
C00044491
|
| 9703 | BCOX, BCOX1, CT101 | KIAA0100 |
C00044491
|
| 9897 | SPG8 | KIAA0196 |
C00044491
|
| 9766 | KIAA0247 |
C00044491
|
|
| 9692 | MRPP3, PRORP | KIAA0391 |
C00044491
|
| 57235 |
C00044491
|
||
| 9764 | KIAA0513 |
C00044491
|
|
| 9851 | KIAA0753 |
C00044491
|
|
| 23313 | C22orf9 | KIAA0930 |
C00044491
|
| 23379 | KIAA0947 |
C00044491
|
|
| 23325 | SWIP | KIAA1033 |
C00044491
|
| 84162 | FSA, Tweek | KIAA1109 |
C00044491
|
| 57456 | KIAA1143 |
C00044491
|
|
| 57189 | LCHN, PRO2561 | KIAA1147 |
C00044491
|
| 57179 | p60MONOX | KIAA1191 |
C00044491
|
| 57214 | CCSP1, TMEM2L | KIAA1199 |
C00044491
|
| 343990 | KIAA1211L, C2orf55 | KIAA1211-like |
C00044491
|
| 57536 | KIAA1328 |
C00044491
|
|
| 57587 | KIAA1430 |
C00044491
|
|
| 57589 | CIP150, bA207C16.1 | KIAA1432 |
C00044491
|
| 57648 | KIAA1522 |
C00044491
|
|
| 57650 | CIP2A, p90 | KIAA1524 |
C00044491
|
| 57698 | KIAA1598 |
C00044491
|
|
| 85379 | KIAA1671 |
C00044491
|
|
| 85459 | KIAA1731 |
C00044491
|
|
| 90231 | KIAA2013 |
C00044491
|
|
| 205717 | KIAA2018 |
C00044491
|
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00044491
|
| 23303 | KIF13B, GAKIN | kinesin family member 13B |
C00044491
|
| 9928 | KIF14 | kinesin family member 14 |
C00044491
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00044491
|
| 55614 | KIF16B, C20orf23, KISC20ORF, SNX23 | kinesin family member 16B |
C00044491
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00044491
|
| 146909 | KIF18B | kinesin family member 18B |
C00044491
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00044491
|
| 9585 | KIF20B, CT90, KRMP1, MPHOSPH1, MPP-1, MPP1 | kinesin family member 20B |
C00044491
|
| 9493 | KIF23, CHO1, KNSL5, MKLP-1, MKLP1 | kinesin family member 23 |
C00044491
|
| 347240 | KIF24, C9orf48, bA571F15.4 | kinesin family member 24 |
C00044491
|
| 11004 | KIF2C, KNSL6, MCAK | kinesin family member 2C |
C00044491
|
| 11127 | KIF3A, FLA10, KLP-20 | kinesin family member 3A |
C00044491
|
| 24137 | KIF4A, KIF4, KIF4G1 | kinesin family member 4A |
C00044491
|
| 221458 | KIF6, C6orf102, dJ1043E3.1, dJ137F1.4, dJ188D3.1 | kinesin family member 6 |
C00044491
|
| 22920 | KIFAP3, FLA3, KAP-1, KAP-3, KAP3, SMAP, Smg-GDS, dJ190I16.1 | kinesin-associated protein 3 |
C00044491
|
| 3833 | KIFC1, HSET, KNSL2 | kinesin family member C1 |
C00044491
|
| 4254 | KITLG, FPH2, KL-1, Kitl, MGF, SCF, SF, SHEP7 | KIT ligand |
C00044491
|
| 64837 | KLC2 | kinesin light chain 2 |
C00044491
|
| 89953 | KLC4, KNSL8, bA387M24.3 | kinesin light chain 4 |
C00044491
|
| 51621 | KLF13, BTEB3, FKLF2, NSLP1, RFLAT-1, RFLAT1 | Kruppel-like factor 13 |
C00044491
|
| 10365 | KLF2, LKLF | Kruppel-like factor 2 |
C00044491
|
| 51274 | KLF3, BKLF | Kruppel-like factor 3 (basic) |
C00044491
|
| 688 | KLF5, BTEB2, CKLF, IKLF | Kruppel-like factor 5 (intestinal) |
C00044491
|
| 1316 | KLF6, BCD1, CBA1, COPEB, CPBP, GBF, PAC1, ST12, ZF9 | Kruppel-like factor 6 |
C00044491
|
| 687 | KLF9, BTEB, BTEB1 | Kruppel-like factor 9 |
C00044491
|
| 23008 | KLHDC10, slim | kelch domain containing 10 |
C00044491
|
| 116138 | KLHDC3, PEAS, RP1-20C7.3, dJ20C7.3 | kelch domain containing 3 |
C00044491
|
| 27252 | KLHL20, KHLHX, KLEIP, KLHLX, RP3-383J4.3 | kelch-like family member 20 |
C00044491
|
| 9903 | KLHL21 | kelch-like family member 21 |
C00044491
|
| 54800 | KLHL24, DRE1, KRIP6 | kelch-like family member 24 |
C00044491
|
| 64410 | KLHL25, ENC-2, ENC2 | kelch-like family member 25 |
C00044491
|
| 54813 | KLHL28, BTBD5 | kelch-like family member 28 |
C00044491
|
| 377007 | KLHL30 | kelch-like family member 30 |
C00044491
|
| 79786 | KLHL36, C16orf44 | kelch-like family member 36 |
C00044491
|
| 57542 | KLHL42, Ctb9, KLHDC5 | kelch-like family member 42 |
C00044491
|
| 51088 | KLHL5 | kelch-like family member 5 |
C00044491
|
| 3824 | KLRD1, CD94 | killer cell lectin-like receptor subfamily D, member 1 |
C00044491
|
| 51348 | KLRF1, CLEC5C, NKp80 | killer cell lectin-like receptor subfamily F, member 1 |
C00044491
|
| 8564 | KMO, dJ317G22.1 | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (EC:1.14.13.9) |
C00044491
|
| 8085 | KMT2D, AAD10, ALR, CAGL114, KABUK1, KMS, MLL2, MLL4, TNRC21 | lysine (K)-specific methyltransferase 2D (EC:2.1.1.43) |
C00044491
|
| 55904 | KMT2E, HDCMC04P, MLL5 | lysine (K)-specific methyltransferase 2E (EC:2.1.1.43) |
C00044491
|
| 148930 | KNCN, Kino, L5 | kinocilin |
C00044491
|
| 90417 | KNSTRN, C15orf23, SKAP, TRAF4AF1 | kinetochore-localized astrin/SPAG5 binding protein |
C00044491
|
| 9735 | KNTC1, ROD | kinetochore associated 1 |
C00044491
|
| 3836 | KPNA1, IPOA5, NPI-1, RCH2, SRP1 | karyopherin alpha 1 (importin alpha 5) |
C00044491
|
| 3840 | KPNA4, IPOA3, QIP1, SRP3 | karyopherin alpha 4 (importin alpha 3) |
C00044491
|
| 3837 | KPNB1, IMB1, IPO1, IPOB, Impnb, NTF97 | karyopherin (importin) beta 1 |
C00044491
|
| 83999 | KREMEN1, KREMEN, KRM1 | kringle containing transmembrane protein 1 |
C00044491
|
| 889 | KRIT1, CAM, CCM1 | KRIT1, ankyrin repeat containing |
C00044491
|
| 3848 | KRT1, CK1, EHK, EHK1, EPPK, K1, KRT1A, NEPPK | keratin 1 |
C00044491
|
| 3859 | KRT12, K12 | keratin 12 |
C00044491
|
| 3866 | KRT15, CK15, K15, K1CO | keratin 15 |
C00044491
|
| 3875 | KRT18, CYK18, K18 | keratin 18 |
C00044491
|
| 3852 | KRT5, CK5, DDD, DDD1, EBS2, K5, KRT5A | keratin 5 |
C00044491
|
| 3856 | KRT8, CARD2, CK-8, CK8, CYK8, K2C8, K8, KO | keratin 8 |
C00044491
|
| 3892 | KRT86, HB6, Hb1, KRTHB1, KRTHB6, MNX, hHb6 | keratin 86 |
C00044491
|
| 100129075 | KTN1-AS1, C14orf33 | KTN1 antisense RNA 1 |
C00044491
|
| 8942 | KYNU | kynureninase (EC:3.7.1.3) |
C00044491
|
| 112849 | L3HYPDH, C14orf149 | L-3-hydroxyproline dehydratase (trans-) (EC:4.2.1.77) |
C00044491
|
| 51110 | LACTB2 | lactamase, beta 2 |
C00044491
|
| 3898 | LAD1, LadA | ladinin 1 |
C00044491
|
| 3904 | LAIR2, CD306 | leukocyte-associated immunoglobulin-like receptor 2 |
C00044491
|
| 3910 | LAMA4, CMD1JJ, LAMA3, LAMA4*-1 | laminin, alpha 4 |
C00044491
|
| 3912 | LAMB1, CLM, LIS5 | laminin, beta 1 |
C00044491
|
| 10319 | LAMC3, OCCM | laminin, gamma 3 |
C00044491
|
| 3920 | LAMP2, CD107b, LAMP-2, LAMPB, LGP110 | lysosomal-associated membrane protein 2 |
C00044491
|
| 27074 | LAMP3, CD208, DC_LAMP, DC-LAMP, DCLAMP, LAMP, LAMP-3, TSC403 | lysosomal-associated membrane protein 3 |
C00044491
|
| 51056 | LAP3, LAP, LAPEP, PEPS | leucine aminopeptidase 3 (EC:3.4.11.5 3.4.11.1) |
C00044491
|
| 23367 | LARP1, LARP | La ribonucleoprotein domain family, member 1 |
C00044491
|
| 113251 | LARP4 | La ribonucleoprotein domain family, member 4 |
C00044491
|
| 55323 | LARP6, ACHN | La ribonucleoprotein domain family, member 6 |
C00044491
|
| 51574 | LARP7, ALAZS, PIP7S | La ribonucleoprotein domain family, member 7 |
C00044491
|
| 23395 | LARS2, LEURS, PRLTS4 | leucyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.4) |
C00044491
|
| 81887 | LAS1L, dJ475B7.2 | LAS1-like (S. cerevisiae) |
C00044491
|
| 3927 | LASP1, Lasp-1, MLN50 | LIM and SH3 protein 1 |
C00044491
|
| 3931 | LCAT | lecithin-cholesterol acyltransferase (EC:2.3.1.43) |
C00044491
|
| 353134 | LCE1D, LEP4 | late cornified envelope 1D |
C00044491
|
| 353142 | LCE3A, LEP13 | late cornified envelope 3A |
C00044491
|
| 353143 | LCE3B, LEP14 | late cornified envelope 3B |
C00044491
|
| 253558 | LCLAT1, 1AGPAT8, AGPAT8, ALCAT1, HSRG1849, LYCAT, UNQ1849 | lysocardiolipin acyltransferase 1 (EC:2.3.1.51) |
C00044491
|
| 51451 | LCMT1, LCMT, PPMT1 | leucine carboxyl methyltransferase 1 (EC:2.1.1.233) |
C00044491
|
| 389812 | LCN15, PRO6093, UNQ2541 | lipocalin 15 |
C00044491
|
| 84458 | LCOR, MLR2 | ligand dependent nuclear receptor corepressor |
C00044491
|
| 3936 | LCP1, CP64, L-PLASTIN, LC64P, LPL, PLS2 | lymphocyte cytosolic protein 1 (L-plastin) |
C00044491
|
| 3937 | LCP2, SLP-76, SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
C00044491
|
| 9079 | LDB2, CLIM1, LDB1 | LIM domain binding 2 |
C00044491
|
| 3939 | LDHA, GSD11, LDH1, LDHM | lactate dehydrogenase A (EC:1.1.1.27) |
C00044491
|
| 3945 | LDHB, LDH-B, LDH-H, LDHBD, TRG-5 | lactate dehydrogenase B (EC:1.1.1.27) |
C00044491
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00044491
|
| 388633 | LDLRAD1 | low density lipoprotein receptor class A domain containing 1 |
C00044491
|
| 26119 | LDLRAP1, ARH, ARH1, ARH2, FHCB1, FHCB2 | low density lipoprotein receptor adaptor protein 1 |
C00044491
|
| 84247 | LDOC1L, Mar6, Mart6, dJ1033E15.2 | leucine zipper, down-regulated in cancer 1-like |
C00044491
|
| 116842 | LEAP2, LEAP-2 | liver expressed antimicrobial peptide 2 |
C00044491
|
| 51176 | LEF1, LEF-1, TCF10, TCF1ALPHA, TCF7L3 | lymphoid enhancer-binding factor 1 |
C00044491
|
| 149018 | LELP1 | late cornified envelope-like proline-rich 1 |
C00044491
|
| 23484 | LEPROTL1, HSPC112, Vps55, my047 | leptin receptor overlapping transcript-like 1 |
C00044491
|
| 3964 | LGALS8, Gal-8, PCTA-1, PCTA1, Po66-CBP | lectin, galactoside-binding, soluble, 8 |
C00044491
|
| 29094 | LGALSL, GRP | lectin, galactoside-binding-like |
C00044491
|
| 163175 | LGI4, LGIL3 | leucine-rich repeat LGI family, member 4 |
C00044491
|
| 5641 | LGMN, AEP, LGMN1, PRSC1 | legumain (EC:3.4.22.34) |
C00044491
|
| 10186 | LHFP | lipoma HMGIC fusion partner (EC:3.6.5.4) |
C00044491
|
| 11019 | LIAS, LAS, LIP1, LS, PDHLD | lipoic acid synthetase (EC:2.8.1.8) |
C00044491
|
| 3978 | LIG1 | ligase I, DNA, ATP-dependent (EC:6.5.1.1) |
C00044491
|
| 3980 | LIG3, LIG2 | ligase III, DNA, ATP-dependent (EC:6.5.1.1) |
C00044491
|
| 10990 | LILRB5, CD85C, LIR-8, LIR8 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 5 |
C00044491
|
| 51474 | LIMA1, EPLIN, SREBP3 | LIM domain and actin binding 1 |
C00044491
|
| 22998 | LIMCH1, LIMCH1A, LMO7B | LIM and calponin homology domains 1 |
C00044491
|
| 8994 | LIMD1 | LIM domains containing 1 |
C00044491
|
| 55957 | LIN37, F25965, ZK418.4, lin-37 | lin-37 homolog (C. elegans) |
C00044491
|
| 91750 | LIN52, C14orf46, c14_5549 | lin-52 homolog (C. elegans) |
C00044491
|
| 283860 | LINC00304, C16orf81, NCRNA00304 | long intergenic non-protein coding RNA 304 |
C00044491
|
| 29092 | LINC00339, HSPC157, NCRNA00339 | long intergenic non-protein coding RNA 339 |
C00044491
|
| 79686 | LINC00341, C14orf139, NCRNA00341 | long intergenic non-protein coding RNA 341 |
C00044491
|
| 84791 | LINC00467, C1orf97, RP11-318L16.3 | long intergenic non-protein coding RNA 467 |
C00044491
|
| 81698 | LINC00597, C15orf5 | long intergenic non-protein coding RNA 597 |
C00044491
|
| 3988 | LIPA, CESD, LAL | lipase A, lysosomal acid, cholesterol esterase (EC:3.1.1.13) |
C00044491
|
| 8513 | LIPF, GL, HGL, HLAL | lipase, gastric (EC:3.1.1.3) |
C00044491
|
| 9388 | LIPG, EDL, EL, PRO719 | lipase, endothelial (EC:3.1.1.3) |
C00044491
|
| 200879 | LIPH, AH, ARWH2, LAH2, LPDLR, PLA1B, mPA-PLA1 | lipase, member H (EC:2.7.11.30) |
C00044491
|
| 64327 | LMBR1, ACHP, C7orf2, DIF14, PPD2, TPT, ZRS | limb development membrane protein 1 |
C00044491
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00044491
|
| 8543 | LMO4 | LIM domain only 4 |
C00044491
|
| 83752 | LONP2, LONP, LONPL | lon peptidase 2, peroxisomal |
C00044491
|
| 10162 | LPCAT3, C3F, LPCAT, LPLAT_5, LPSAT, MBOAT5, OACT5, nessy | lysophosphatidylcholine acyltransferase 3 (EC:2.3.1.23 2.3.1.n6) |
C00044491
|
| 9926 | LPGAT1, FAM34A, FAM34A1, NET8 | lysophosphatidylglycerol acyltransferase 1 |
C00044491
|
| 22859 | LPHN1, CIRL1, CL1, LEC2 | latrophilin 1 |
C00044491
|
| 23266 | LPHN2, CIRL2, CL2, LEC1, LPHH1 | latrophilin 2 |
C00044491
|
| 9663 | LPIN2 | lipin 2 (EC:3.1.3.4) |
C00044491
|
| 4026 | LPP | LIM domain containing preferred translocation partner in lipoma |
C00044491
|
| 64748 | LPPR2, PRG4 | lipid phosphate phosphatase-related protein type 2 (EC:3.1.3.4) |
C00044491
|
| 9404 | LPXN, LDPL | leupaxin |
C00044491
|
| 987 | LRBA, BGL, CDC4L, CVID8, LAB300, LBA | LPS-responsive vesicle trafficking, beach and anchor containing |
C00044491
|
| 57913 |
C00044491
|
||
| 116844 | LRG1, HMFT1766, LRG | leucine-rich alpha-2-glycoprotein 1 |
C00044491
|
| 55791 | LRIF1, C1orf103, RIF1, RP11-96K19.1 | ligand dependent nuclear receptor interacting factor 1 |
C00044491
|
| 26020 | LRP10, LRP9, MST087 | low density lipoprotein receptor-related protein 10 |
C00044491
|
| 84918 | LRP11, MANSC3, bA350J20.3 | low density lipoprotein receptor-related protein 11 |
C00044491
|
| 4038 | LRP4, CLSS, LRP-4, LRP10, MEGF7, SOST2 | low density lipoprotein receptor-related protein 4 |
C00044491
|
| 4041 | LRP5, BMND1, EVR1, EVR4, HBM, LR3, LRP7, OPPG, OPS, OPTA1, VBCH2 | low density lipoprotein receptor-related protein 5 |
C00044491
|
| 4043 | LRPAP1, A2MRAP, A2RAP, HBP44, MRAP, MYP23, RAP | low density lipoprotein receptor-related protein associated protein 1 |
C00044491
|
| 9684 | LRRC14, LRRC14A | leucine rich repeat containing 14 |
C00044491
|
| 55604 | LRRC16A, CARMIL, CARMIL1, CARMIL1a, LRRC16, dJ501N12.1, dJ501N12.5 | leucine rich repeat containing 16A |
C00044491
|
| 55631 | LRRC40, dJ677H15.1 | leucine rich repeat containing 40 |
C00044491
|
| 149499 | LRRC71, C1orf92, RP11-356J7.1 | leucine rich repeat containing 71 |
C00044491
|
| 55144 | LRRC8D, LRRC5 | leucine rich repeat containing 8 family, member D |
C00044491
|
| 85444 | LRRCC1, CLERC, SAP2, VFL1 | leucine rich repeat and coiled-coil centrosomal protein 1 |
C00044491
|
| 9208 | LRRFIP1, FLAP-1, FLAP1, FLIIAP1, GCF-2, GCF2, HUFI-1, TRIP | leucine rich repeat (in FLII) interacting protein 1 |
C00044491
|
| 9209 | LRRFIP2, HUFI-2 | leucine rich repeat (in FLII) interacting protein 2 |
C00044491
|
| 10446 | LRRN2, FIGLER7, GAC1, LRANK1, LRRN5 | leucine rich repeat neuronal 2 |
C00044491
|
| 26065 | LSM14A, C19orf13, FAM61A, RAP55, RAP55A | LSM14A, SCD6 homolog A (S. cerevisiae) |
C00044491
|
| 286006 | LSMEM1, C7orf53 | leucine-rich single-pass membrane protein 1 |
C00044491
|
| 4047 | LSS, OSC | lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (EC:5.4.99.7) |
C00044491
|
| 4049 | LTA, LT, TNFB, TNFSF1 | lymphotoxin alpha |
C00044491
|
| 4048 | LTA4H | leukotriene A4 hydrolase (EC:3.3.2.6) |
C00044491
|
| 26046 | LTN1, C21orf10, C21orf98, RNF160, ZNF294 | listerin E3 ubiquitin protein ligase 1 |
C00044491
|
| 7798 | LUZP1, LUZP | leucine zipper protein 1 |
C00044491
|
| 51213 | LUZP4, CT-28, CT-8, CT28, HOM-TES-85 | leucine zipper protein 4 |
C00044491
|
| 56925 | LXN, ECI, TCI | latexin |
C00044491
|
| 8581 | LY6D, E48, Ly-6D | lymphocyte antigen 6 complex, locus D |
C00044491
|
| 80740 | LY6G6C, C6orf24, G6c, NG24 | lymphocyte antigen 6 complex, locus G6C |
C00044491
|
| 23643 | LY96, ESOP-1, MD-2, MD2, ly-96 | lymphocyte antigen 96 |
C00044491
|
| 55646 | LYAR, ZC2HC2, ZLYAR | Ly1 antibody reactive |
C00044491
|
| 129530 | LYG1, SALW1939 | lysozyme G-like 1 |
C00044491
|
| 127018 | LYPLAL1 | lysophospholipase-like 1 (EC:3.1.2.-) |
C00044491
|
| 10894 | LYVE1, CRSBP-1, HAR, LYVE-1, XLKD1 | lymphatic vessel endothelial hyaluronan receptor 1 |
C00044491
|
| 9762 | LZTS3, PROSAPIP1 | leucine zipper, putative tumor suppressor family member 3 |
C00044491
|
| 4074 | M6PR, CD-MPR, MPR_46, MPR-46, MPR46, SMPR | mannose-6-phosphate receptor (cation dependent) |
C00044491
|
| 23499 | MACF1, ABP620, ACF7, MACF, OFC4 | microtubule-actin crosslinking factor 1 |
C00044491
|
| 28992 | MACROD1, LRP16 | MACRO domain containing 1 |
C00044491
|
| 9935 | MAFB, KRML, MCTO | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
C00044491
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00044491
|
| 64110 | MAGEF1 | melanoma antigen family F, 1 |
C00044491
|
| 9223 | MAGI1, AIP-3, AIP3, BAIAP1, BAP-1, BAP1, MAGI-1, TNRC19, WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 |
C00044491
|
| 9863 | MAGI2, ACVRIP1, AIP-1, AIP1, ARIP1, MAGI-2, SSCAM | membrane associated guanylate kinase, WW and PDZ domain containing 2 |
C00044491
|
| 4117 | MAK, RP62, dJ417M14.2 | male germ cell-associated kinase (EC:2.7.11.22) |
C00044491
|
| 4118 | MAL | mal, T-cell differentiation protein |
C00044491
|
| 114569 | MAL2 | mal, T-cell differentiation protein 2 (gene/pseudogene) |
C00044491
|
| 10905 | MAN1A2, MAN1B | mannosidase, alpha, class 1A, member 2 (EC:3.2.1.113) |
C00044491
|
| 4124 | MAN2A1, GOLIM7, MANA2, MANII | mannosidase, alpha, class 2A, member 1 (EC:3.2.1.114) |
C00044491
|
| 23324 | MAN2B2 | mannosidase, alpha, class 2B, member 2 (EC:3.2.1.24) |
C00044491
|
| 4126 | MANBA, MANB1 | mannosidase, beta A, lysosomal (EC:3.2.1.25) |
C00044491
|
| 54682 | MANSC1, 9130403P13Rik, LOH12CR3 | MANSC domain containing 1 (EC:3.2.1.14) |
C00044491
|
| 4128 | MAOA, MAO-A | monoamine oxidase A (EC:1.4.3.4) |
C00044491
|
| 4131 | MAP1B, FUTSCH, MAP5 | microtubule-associated protein 1B |
C00044491
|
| 81631 | MAP1LC3B, ATG8F, LC3B, MAP1A/1BLC3, MAP1LC3B-a | microtubule-associated protein 1 light chain 3 beta |
C00044491
|
| 55201 | MAP1S, BPY2IP1, C19orf5, MAP8, VCY2IP-1, VCY2IP1 | microtubule-associated protein 1S |
C00044491
|
| 5607 | MAP2K5, HsT17454, MAPKK5, MEK5, PRKMK5 | mitogen-activated protein kinase kinase 5 (EC:2.7.12.2) |
C00044491
|
| 5609 | MAP2K7, Jnkk2, MAPKK7, MKK7, PRKMK7, SAPKK-4, SAPKK4 | mitogen-activated protein kinase kinase 7 (EC:2.7.12.2) |
C00044491
|
| 7786 | MAP3K12, DLK, MEKK12, MUK, ZPK, ZPKP1 | mitogen-activated protein kinase kinase kinase 12 (EC:2.7.11.25) |
C00044491
|
| 9175 | MAP3K13, LZK, MEKK13, MLK | mitogen-activated protein kinase kinase kinase 13 (EC:2.7.11.25) |
C00044491
|
| 9020 | MAP3K14, FTDCR1B, HS, HSNIK, NIK | mitogen-activated protein kinase kinase kinase 14 (EC:2.7.11.25) |
C00044491
|
| 4215 | MAP3K3, MAPKKK3, MEKK3 | mitogen-activated protein kinase kinase kinase 3 (EC:2.7.11.25) |
C00044491
|
| 4217 | MAP3K5, ASK1, MAPKKK5, MEKK5 | mitogen-activated protein kinase kinase kinase 5 (EC:2.7.11.25) |
C00044491
|
| 4134 | MAP4 | microtubule-associated protein 4 |
C00044491
|
| 9053 | MAP7, E-MAP-115, EMAP115 | microtubule-associated protein 7 |
C00044491
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00044491
|
| 5603 | MAPK13, MAPK_13, MAPK-13, PRKM13, SAPK4, p38delta | mitogen-activated protein kinase 13 (EC:2.7.11.24) |
C00044491
|
| 1432 | MAPK14, CSBP, CSBP1, CSBP2, CSPB1, EXIP, Mxi2, PRKM14, PRKM15, RK, SAPK2A, p38, p38ALPHA | mitogen-activated protein kinase 14 (EC:2.7.11.24) |
C00044491
|
| 93487 | MAPK1IP1L, C14orf32, MISS, c14_5346 | mitogen-activated protein kinase 1 interacting protein 1-like |
C00044491
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00044491
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00044491
|
| 23162 | MAPK8IP3, JIP3, JSAP1, SYD2, syd | mitogen-activated protein kinase 8 interacting protein 3 |
C00044491
|
| 5601 | MAPK9, JNK-55, JNK2, JNK2A, JNK2ALPHA, JNK2B, JNK2BETA, PRKM9, SAPK, SAPK1a, p54a, p54aSAPK | mitogen-activated protein kinase 9 (EC:2.7.11.24) |
C00044491
|
| 79109 | MAPKAP1, JC310, MIP1, SIN1, SIN1b, SIN1g | mitogen-activated protein kinase associated protein 1 |
C00044491
|
| 7867 | MAPKAPK3, 3PK, MAPKAP-K3, MAPKAP3, MAPKAPK-3, MK-3 | mitogen-activated protein kinase-activated protein kinase 3 (EC:2.7.11.1) |
C00044491
|
| 22919 | MAPRE1, EB1 | microtubule-associated protein, RP/EB family, member 1 |
C00044491
|
| 22924 | MAPRE3, EB3, EBF3, EBF3-S, RP3 | microtubule-associated protein, RP/EB family, member 3 |
C00044491
|
| 51257 | MARCH2, MARCH-II, RNF172 | membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00044491
|
| 65108 | MARCKSL1, F52, MACMARCKS, MLP, MLP1, MRP | MARCKS-like 1 |
C00044491
|
| 57787 | MARK4, MARK4L, MARK4S, MARKL1, MARKL1L, PAR-1D | MAP/microtubule affinity-regulating kinase 4 (EC:2.7.11.1) |
C00044491
|
| 153562 | MARVELD2, DFNB49, MARVD2, MRVLDC2, Tric | MARVEL domain containing 2 |
C00044491
|
| 5648 | MASP1, 3MC1, CRARF, CRARF1, MAP1, MASP, MASP3, MAp44, PRSS5, RaRF | mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) (EC:3.4.21.-) |
C00044491
|
| 23031 | MAST3 | microtubule associated serine/threonine kinase 3 (EC:2.7.11.1) |
C00044491
|
| 84930 | MASTL, GREATWALL, GW, GWL, MAST-L, RP11-85G18.2, THC2, hGWL | microtubule associated serine/threonine kinase-like (EC:2.7.11.1) |
C00044491
|
| 57506 | MAVS, CARDIF, IPS-1, IPS1, VISA | mitochondrial antiviral signaling protein |
C00044491
|
| 8930 | MBD4, MED1 | methyl-CpG binding domain protein 4 |
C00044491
|
| 55777 | MBD5, MRD1 | methyl-CpG binding domain protein 5 |
C00044491
|
| 4154 | MBNL1, EXP, EXP35, EXP40, EXP42, MBNL | muscleblind-like splicing regulator 1 |
C00044491
|
| 10150 | MBNL2, MBLL, MBLL39, PRO2032 | muscleblind-like splicing regulator 2 |
C00044491
|
| 55796 | MBNL3, CHCR, MBLX, MBLX39, MBXL | muscleblind-like splicing regulator 3 |
C00044491
|
| 54799 | MBTD1, SA49P01 | mbt domain containing 1 |
C00044491
|
| 4162 | MCAM, CD146, MUC18 | melanoma cell adhesion molecule |
C00044491
|
| 27349 | MCAT, FASN2C, MCT, MT, NET62, fabD | malonyl CoA:ACP acyltransferase (mitochondrial) (EC:2.3.1.39) |
C00044491
|
| 4163 | MCC, MCC1 | mutated in colorectal cancers |
C00044491
|
| 23101 | MCF2L2, ARHGEF22 | MCF.2 cell line derived transforming sequence-like 2 |
C00044491
|
| 90411 | MCFD2, F5F8D, F5F8D2, LMAN1IP, SDNSF | multiple coagulation factor deficiency 2 |
C00044491
|
| 4170 | MCL1, BCL2L3, EAT, MCL1-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT | myeloid cell leukemia sequence 1 (BCL2-related) |
C00044491
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00044491
|
| 4172 | MCM3, HCC5, P1-MCM3, P1.h, RLFB | minichromosome maintenance complex component 3 (EC:3.6.4.12) |
C00044491
|
| 8888 | MCM3AP, GANP, MAP80, SAC3 | minichromosome maintenance complex component 3 associated protein |
C00044491
|
| 4173 | MCM4, CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 | minichromosome maintenance complex component 4 (EC:3.6.4.12) |
C00044491
|
| 4176 | MCM7, CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146 | minichromosome maintenance complex component 7 (EC:3.6.4.12) |
C00044491
|
| 84515 | MCM8, C20orf154, dJ967N21.5 | minichromosome maintenance complex component 8 (EC:3.6.4.12) |
C00044491
|
| 57192 | MCOLN1, MG-2, ML4, MLIV, MST080, TRP-ML1, TRPM-L1, TRPML1 | mucolipin 1 |
C00044491
|
| 90550 | MCU, C10orf42, CCDC109A | mitochondrial calcium uniporter |
C00044491
|
| 9656 | MDC1, NFBD1 | mediator of DNA-damage checkpoint 1 |
C00044491
|
| 4193 | MDM2, ACTFS, HDMX, hdm2 | MDM2 oncogene, E3 ubiquitin protein ligase |
C00044491
|
| 23195 | MDN1 | MDN1, midasin homolog (yeast) |
C00044491
|
| 64769 | MEAF6, C1orf149, CENP-28, EAF6, NY-SAR-91 | MYST/Esa1-associated factor 6 |
C00044491
|
| 2122 | MECOM, AML1-EVI-1, EVI1, MDS1, MDS1-EVI1, PRDM3 | MDS1 and EVI1 complex locus |
C00044491
|
| 9968 | MED12, ARC240, CAGH45, FGS1, HOPA, MED12S, OHDOX, OKS, OPA1, TNRC11, TRAP230 | mediator complex subunit 12 |
C00044491
|
| 9412 | MED21, SRB7, SURB7, hSrb7 | mediator complex subunit 21 |
C00044491
|
| 81857 | MED25, ACID1, ARC92, CMT2B2, P78, PTOV2 | mediator complex subunit 25 |
C00044491
|
| 9441 | MED26, CRSP7, CRSP70 | mediator complex subunit 26 |
C00044491
|
| 80306 | MED28, 1500003D12Rik, EG1, magicin | mediator complex subunit 28 |
C00044491
|
| 55588 | MED29, IXL | mediator complex subunit 29 |
C00044491
|
| 51003 | MED31, 3110004H13Rik, Soh1 | mediator complex subunit 31 |
C00044491
|
| 10001 | MED6, ARC33, NY-REN-28 | mediator complex subunit 6 |
C00044491
|
| 1955 | MEGF9, EGFL5 | multiple EGF-like-domains 9 |
C00044491
|
| 4212 | MEIS2, HsT18361, MRG1 | Meis homeobox 2 |
C00044491
|
| 9833 | MELK, HPK38 | maternal embryonic leucine zipper kinase (EC:2.7.11.1 2.7.10.2) |
C00044491
|
| 4223 | MEOX2, GAX, MOX2 | mesenchyme homeobox 2 |
C00044491
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00044491
|
| 10988 | METAP2, MAP2, MNPEP, p67, p67eIF2 | methionyl aminopeptidase 2 (EC:3.4.11.18) |
C00044491
|
| 284207 | METRNL | meteorin, glial cell differentiation regulator-like |
C00044491
|
| 399818 | METTL10, C10orf138, Em:AC068896.3 | methyltransferase like 10 |
C00044491
|
| 751071 | METTL12, U99HG | methyltransferase like 12 (EC:2.1.1.-) |
C00044491
|
| 51603 | METTL13, 5630401D24Rik, KIAA0859, feat | methyltransferase like 13 |
C00044491
|
| 92342 | METTL18, AsTP2, C1orf156, HPM1 | methyltransferase like 18 |
C00044491
|
| 124512 | METTL23, C17orf95 | methyltransferase like 23 |
C00044491
|
| 56339 | METTL3, IME4, M6A, MT-A70, Spo8 | methyltransferase like 3 (EC:2.1.1.62) |
C00044491
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00044491
|
| 4237 | MFAP2, MAGP, MAGP-1, MAGP1 | microfibrillar-associated protein 2 |
C00044491
|
| 56947 | MFF, C2orf33, GL004 | mitochondrial fission factor |
C00044491
|
| 4240 | MFGE8, BA46, EDIL1, HMFG, HsT19888, MFG-E8, MFGM, OAcGD3S, SED1, SPAG10, hP47 | milk fat globule-EGF factor 8 protein |
C00044491
|
| 9258 | MFHAS1, LRRC65, MASL1 | malignant fibrous histiocytoma amplified sequence 1 |
C00044491
|
| 4241 | MFI2, CD228, MAP97, MTF1 | antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
C00044491
|
| 55669 | MFN1, hfzo1, hfzo2 | mitofusin 1 |
C00044491
|
| 9927 | MFN2, CMT2A, CMT2A2, CPRP1, HSG, MARF | mitofusin 2 |
C00044491
|
| 79157 | MFSD11, ET | major facilitator superfamily domain containing 11 |
C00044491
|
| 126321 | MFSD12, C19orf28, PP3501 | major facilitator superfamily domain containing 12 |
C00044491
|
| 84975 | MFSD5 | major facilitator superfamily domain containing 5 |
C00044491
|
| 11282 | MGAT4B, GNT-IV, GNT-IVB | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (EC:2.4.1.145) |
C00044491
|
| 10724 | MGEA5, MEA5, NCOAT, OGA | meningioma expressed antigen 5 (hyaluronidase) (EC:2.3.1.48 3.2.1.169) |
C00044491
|
| 11343 | MGLL, HU-K5, HUK5, MAGL, MGL | monoglyceride lipase (EC:3.1.1.23) |
C00044491
|
| 92667 | MGME1, C20orf72, DDK1, MTDPS11, bA504H3.4 | mitochondrial genome maintenance exonuclease 1 |
C00044491
|
| 4257 | MGST1, GST12, MGST, MGST-I | microsomal glutathione S-transferase 1 (EC:2.5.1.18) |
C00044491
|
| 4258 | MGST2, GST2, MGST-II | microsomal glutathione S-transferase 2 (EC:2.5.1.18) |
C00044491
|
| 57534 | MIB1, DIP-1, DIP1, LVNC7, MIB, ZZANK2, ZZZ6 | mindbomb E3 ubiquitin protein ligase 1 |
C00044491
|
| 142678 | MIB2, ZZANK1, ZZZ5 | mindbomb E3 ubiquitin protein ligase 2 |
C00044491
|
| 100507436 | MICA, MIC-A, PERB11.1 | MHC class I polypeptide-related sequence A |
C00044491
|
| 9645 | MICAL2, MICAL-2, MICAL2PV1, MICAL2PV2 | microtubule associated monooxygenase, calponin and LIM domain containing 2 |
C00044491
|
| 57553 | MICAL3, MICAL-3 | microtubule associated monooxygenase, calponin and LIM domain containing 3 |
C00044491
|
| 85377 | MICALL1, MICAL-L1, MIRAB13 | MICAL-like 1 |
C00044491
|
| 4277 | MICB, PERB11.2 | MHC class I polypeptide-related sequence B |
C00044491
|
| 90007 | MIDN | midnolin |
C00044491
|
| 54471 | MIEF1, HSU79252, MID51, SMCR7L, dJ1104E15.3 | mitochondrial elongation factor 1 |
C00044491
|
| 84864 | MINA, MDIG, MINA53, NO52, ROX | MYC induced nuclear antigen |
C00044491
|
| 50488 | MINK1, B55, MAP4K6, MINK, YSK2, ZC3, hMINK, hMINKbeta | misshapen-like kinase 1 (EC:2.7.11.1) |
C00044491
|
| 440574 | MINOS1, C1orf151, MIO10 | mitochondrial inner membrane organizing system 1 |
C00044491
|
| 406913 | MIR126, MIRN126, miRNA126 | microRNA 126 |
C00044491
|
| 406979 | MIR19A, MIRN19A, hsa-mir-19a, miR-19a, miRNA19A | microRNA 19a |
C00044491
|
| 407026 | MIR29C, MIRN29C, miRNA29C | microRNA 29c |
C00044491
|
| 407040 | MIR34A, MIRN34A, miRNA34A, mir-34 | microRNA 34a |
C00044491
|
| 442919 | MIR374A, MIRN374, MIRN374A, hsa-mir-374 | microRNA 374a |
C00044491
|
| 574508 | MIR505, MIRN505, hsa-mir-505 | microRNA 505 |
C00044491
|
| 81571 | MIR600HG, C9orf45, NCRNA00287 | MIR600 host gene (non-protein coding) |
C00044491
|
| 768219 | MIR802, MIRN802, hsa-mir-802 | microRNA 802 |
C00044491
|
| 129531 | MITD1 | MIT, microtubule interacting and transport, domain containing 1 |
C00044491
|
| 4288 | MKI67, KIA, MIB-1 | marker of proliferation Ki-67 |
C00044491
|
| 57591 | MKL1, BSAC, MAL, MRTF-A | megakaryoblastic leukemia (translocation) 1 |
C00044491
|
| 57496 | MKL2, MRTF-B, NPD001 | MKL/myocardin-like 2 |
C00044491
|
| 4289 | MKLN1, TWA2 | muskelin 1, intracellular mediator containing kelch motifs |
C00044491
|
| 8569 | MKNK1, MNK1 | MAP kinase interacting serine/threonine kinase 1 (EC:2.7.11.1) |
C00044491
|
| 2872 | MKNK2, GPRK7, MNK2 | MAP kinase interacting serine/threonine kinase 2 (EC:2.7.11.1) |
C00044491
|
| 9761 | MLEC, KIAA0152 | malectin |
C00044491
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00044491
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00044491
|
| 197259 | MLKL | mixed lineage kinase domain-like |
C00044491
|
| 8028 | MLLT10, AF10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
C00044491
|
| 653483 | MLLT4-AS1, C6orf124, HGC6.4, dJ431P23.3 | MLLT4 antisense RNA 1 (head to head) |
C00044491
|
| 4302 | MLLT6, AF17 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
C00044491
|
| 22877 | MLXIP, MIR, MONDOA, bHLHe36 | MLX interacting protein |
C00044491
|
| 166785 | MMAA, cblA | methylmalonic aciduria (cobalamin deficiency) cblA type |
C00044491
|
| 25974 | MMACHC, RP11-291L19.3, cblC | methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
C00044491
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00044491
|
| 4319 | MMP10, SL-2, STMY2 | matrix metallopeptidase 10 (stromelysin 2) (EC:3.4.24.22) |
C00044491
|
| 4326 | MMP17, MT4-MMP | matrix metallopeptidase 17 (membrane-inserted) (EC:3.4.24.-) |
C00044491
|
| 79812 | MMRN2, EMILIN-3, EMILIN3, ENDOGLYX-1 | multimerin 2 |
C00044491
|
| 84057 | MND1, GAJ | meiotic nuclear divisions 1 homolog (S. cerevisiae) |
C00044491
|
| 64112 | MOAP1, MAP-1, PNMA4 | modulator of apoptosis 1 |
C00044491
|
| 25843 | MOB4, 2C4D, MOB1, MOB3, MOBKL3, PHOCN, PREI3 | MOB family member 4, phocein |
C00044491
|
| 712156 |
C00044491
|
||
| 80168 | MOGAT2, DGAT2L5, MGAT2 | monoacylglycerol O-acyltransferase 2 (EC:2.3.1.22) |
C00044491
|
| 84315 | MON1A, SAND1 | MON1 secretory trafficking family member A |
C00044491
|
| 22880 | MORC2, ZCW3, ZCWCC1 | MORC family CW-type zinc finger 2 |
C00044491
|
| 150291 | MORC2-AS1, C22orf27, NCRNA00325 | MORC2 antisense RNA 1 |
C00044491
|
| 79710 | MORC4, ZCW4, ZCWCC2, dJ75H8.2 | MORC family CW-type zinc finger 4 |
C00044491
|
| 10934 |
C00044491
|
||
| 9643 | MORF4L2, MORFL2, MRGX | mortality factor 4 like 2 |
C00044491
|
| 56180 | MOSPD1, DJ473B4 | motile sperm domain containing 1 |
C00044491
|
| 158747 | MOSPD2 | motile sperm domain containing 2 |
C00044491
|
| 8777 | MPDZ, HYC2, MUPP1 | multiple PDZ domain protein |
C00044491
|
| 10199 | MPHOSPH10, CT90, MPP10, MPP10P | M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
C00044491
|
| 10200 | MPHOSPH6, MPP, MPP-6, MPP6 | M-phase phosphoprotein 6 |
C00044491
|
| 54737 | MPHOSPH8, HSMPP8, TWA3, mpp8 | M-phase phosphoprotein 8 (EC:2.4.2.30) |
C00044491
|
| 10198 | MPHOSPH9, MPP-9, MPP9 | M-phase phosphoprotein 9 |
C00044491
|
| 4354 | MPP1, AAG12, DXS552E, EMP55, MRG1, PEMP | membrane protein, palmitoylated 1, 55kDa |
C00044491
|
| 23164 | MPRIP, M-RIP, MRIP, RHOIP3, RIP3, p116Rip | myosin phosphatase Rho interacting protein |
C00044491
|
| 4357 | MPST, MST, TST2 | mercaptopyruvate sulfurtransferase (EC:2.8.1.2) |
C00044491
|
| 4358 | MPV17, MTDPS6, SYM1 | MpV17 mitochondrial inner membrane protein |
C00044491
|
| 84769 | MPV17L2, FKSG24 | MPV17 mitochondrial membrane protein-like 2 |
C00044491
|
| 3140 | MR1, HLALS | major histocompatibility complex, class I-related |
C00044491
|
| 4361 | MRE11A, ATLD, HNGS1, MRE11, MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
C00044491
|
| 80133 | MROH9, ARMC11, C1orf129 | maestro heat-like repeat family member 9 |
C00044491
|
| 29088 | MRPL15, L15mt, MRP-L15, MRP-L7, RPML7 | mitochondrial ribosomal protein L15 |
C00044491
|
| 54948 | MRPL16, L16mt, MRP-L16 | mitochondrial ribosomal protein L16 |
C00044491
|
| 29074 | MRPL18, L18mt, MRP-L18 | mitochondrial ribosomal protein L18 |
C00044491
|
| 29093 | MRPL22, L22mt, MRP-L22, MRP-L25, RPML25 | mitochondrial ribosomal protein L22 |
C00044491
|
| 6150 | MRPL23, L23MRP, RPL23, RPL23L | mitochondrial ribosomal protein L23 |
C00044491
|
| 54148 | MRPL39, C21orf92, L39mt, MRP-L5, MRPL5, PRED22, PRED66, RPML5 | mitochondrial ribosomal protein L39 |
C00044491
|
| 51073 | MRPL4, CGI-28, L4mt | mitochondrial ribosomal protein L4 |
C00044491
|
| 64975 | MRPL41, BMRP, MRP-L27, MRPL27, RPML27 | mitochondrial ribosomal protein L41 |
C00044491
|
| 84545 | MRPL43, bMRP36a | mitochondrial ribosomal protein L43 |
C00044491
|
| 26589 | MRPL46, C15orf4, LIECG2, P2ECSL | mitochondrial ribosomal protein L46 |
C00044491
|
| 64963 | MRPS11, HCC-2 | mitochondrial ribosomal protein S11 |
C00044491
|
| 28973 | MRPS18B, C6orf14, HSPC183, HumanS18a, MRP-S18-2, MRPS18-2, PTD017, S18amt | mitochondrial ribosomal protein S18B |
C00044491
|
| 10240 | MRPS31, IMOGN38, MRP-S31, S31mt | mitochondrial ribosomal protein S31 |
C00044491
|
| 65993 | MRPS34, MRP-S12, MRP-S34, MRPS12 | mitochondrial ribosomal protein S34 |
C00044491
|
| 64968 | MRPS6, C21orf101, MRP-S6, RPMS6, S6mt | mitochondrial ribosomal protein S6 |
C00044491
|
| 51338 | MS4A4A, 4SPAN1, CD20-L1, CD20L1, MS4A4, MS4A7 | membrane-spanning 4-domains, subfamily A, member 4A |
C00044491
|
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00044491
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00044491
|
| 124540 | MSI2, MSI2H | musashi RNA-binding protein 2 |
C00044491
|
| 55167 | MSL2, MSL-2, MSL2L1, RNF184 | male-specific lethal 2 homolog (Drosophila) |
C00044491
|
| 4477 | MSMB, HPC13, IGBF, MSP, MSPB, PN44, PRPS, PSP, PSP-94, PSP57, PSP94 | microseminoprotein, beta- |
C00044491
|
| 4478 | MSN | moesin |
C00044491
|
| 51734 | MSRB1, SELR, SELX, SEPX1, SepR | methionine sulfoxide reductase B1 (EC:1.8.4.-) |
C00044491
|
| 51765 | MST4, MASK | serine/threonine protein kinase MST4 (EC:2.7.11.1) |
C00044491
|
| 55154 | MSTO1, MST | misato 1, mitochondrial distribution and morphology regulator |
C00044491
|
| 4487 | MSX1, ECTD3, HOX7, HYD1, STHAG1 | msh homeobox 1 |
C00044491
|
| 55545 | MSX2P1, HPX5, HSHPX5, MSX2P | msh homeobox 2 pseudogene 1 |
C00044491
|
| 4490 | MT1B, MT1, MT1Q, MTP | metallothionein 1B |
C00044491
|
| 4495 | MT1G, MT1, MT1K | metallothionein 1G |
C00044491
|
| 4496 | MT1H, MT-0, MT-1H, MT-IH, MT1 | metallothionein 1H |
C00044491
|
| 4499 | MT1M, MT-1M, MT-IM, MT1, MT1K | metallothionein 1M |
C00044491
|
| 4501 | MT1X, MT-1l, MT1 | metallothionein 1X |
C00044491
|
| 4507 | MTAP, BDMF, DMSFH, DMSMFH, LGMBF, MSAP, c86fus | methylthioadenosine phosphorylase (EC:2.4.2.28) |
C00044491
|
| 27085 | MTBP, MDM2BP | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
C00044491
|
| 23787 | MTCH1, CGI-64, PIG60, PSAP, SLC25A49 | mitochondrial carrier 1 |
C00044491
|
| 92140 | MTDH, 3D3, AEG-1, AEG1, LYRIC, LYRIC/3D3 | metadherin |
C00044491
|
| 51001 | MTERFD1, mTERF3 | MTERF domain containing 1 |
C00044491
|
| 130916 | MTERFD2, MTERF4 | MTERF domain containing 2 |
C00044491
|
| 80298 | MTERFD3, mTERF2, mTERFL | MTERF domain containing 3 |
C00044491
|
| 113115 | MTFR2, DUFD1, FAM54A | mitochondrial fission regulator 2 |
C00044491
|
| 25902 | MTHFD1L, FTHFSDC1, MTC1THFS, dJ292B18.2 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like (EC:6.3.4.3) |
C00044491
|
| 10797 | MTHFD2, NMDMC | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase (EC:3.5.4.9 1.5.1.15) |
C00044491
|
| 4524 | MTHFR | methylenetetrahydrofolate reductase (NAD(P)H) (EC:1.5.1.20) |
C00044491
|
| 4528 | MTIF2 | mitochondrial translational initiation factor 2 |
C00044491
|
| 8776 | MTMR1 | myotubularin related protein 1 |
C00044491
|
| 54893 | MTMR10 | myotubularin related protein 10 |
C00044491
|
| 10903 | MTMR11, CRA, RP11-212K13.1 | myotubularin related protein 11 |
C00044491
|
| 9110 | MTMR4, FYVE-DSP2, ZFYVE11 | myotubularin related protein 4 (EC:3.1.3.48) |
C00044491
|
| 66036 | MTMR9, C8orf9, LIP-STYX, MTMR8 | myotubularin related protein 9 |
C00044491
|
| 25821 | MTO1, COXPD10 | mitochondrial tRNA translation optimization 1 |
C00044491
|
| 4548 | MTR, HMAG, MS, cblG | 5-methyltetrahydrofolate-homocysteine methyltransferase (EC:2.1.1.13) |
C00044491
|
| 9788 | MTSS1, MIM, MIMA, MIMB | metastasis suppressor 1 |
C00044491
|
| 4547 | MTTP, ABL, MTP | microsomal triglyceride transfer protein |
C00044491
|
| 57509 | MTUS1, ATBP, ATIP, ICIS, MP44, MTSG1 | microtubule associated tumor suppressor 1 |
C00044491
|
| 56667 | MUC13, DRCC1, MUC-13 | mucin 13, cell surface associated |
C00044491
|
| 200958 | MUC20, MUC-20 | mucin 20, cell surface associated |
C00044491
|
| 394263 | MUC21, C6orf205, KMQK697, MUC-21, bCX31G15.2 | mucin 21, cell surface associated |
C00044491
|
| 79594 | MUL1, C1orf166, GIDE, MAPL, MULAN, RNF218, RP11-401M16.2 | mitochondrial E3 ubiquitin protein ligase 1 |
C00044491
|
| 84939 | MUM1, EXPAND1, HSPC211, MUM-1 | melanoma associated antigen (mutated) 1 |
C00044491
|
| 9961 | MVP, LRP, VAULT1 | major vault protein |
C00044491
|
| 4599 | MX1, IFI-78K, IFI78, MX, MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
C00044491
|
| 4084 | MXD1, BHLHC58, MAD, MAD1 | MAX dimerization protein 1 |
C00044491
|
| 54587 | MXRA8 | matrix-remodelling associated 8 |
C00044491
|
| 4603 | MYBL1, A-MYB, AMYB | v-myb avian myeloblastosis viral oncogene homolog-like 1 |
C00044491
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00044491
|
| 4615 | MYD88, MYD88D | myeloid differentiation primary response 88 |
C00044491
|
| 4619 | MYH1, MYHSA1, MYHa, MyHC-2X/D, MyHC-2x | myosin, heavy chain 1, skeletal muscle, adult |
C00044491
|
| 4621 | MYH3, HEMHC, MYHC-EMB, MYHSE1, SMHCE | myosin, heavy chain 3, skeletal muscle, embryonic |
C00044491
|
| 10398 | MYL9, LC20, MLC-2C, MLC2, MRLC1, MYRL2 | myosin, light chain 9, regulatory |
C00044491
|
| 29116 | MYLIP, IDOL, MIR | myosin regulatory light chain interacting protein (EC:6.3.2.-) |
C00044491
|
| 4638 | MYLK, AAT7, KRP, MLCK, MLCK1, MLCK108, MLCK210, MSTP083, MYLK1, smMLCK | myosin light chain kinase (EC:2.7.11.18) |
C00044491
|
| 340156 | MYLK4, SgK085 | myosin light chain kinase family, member 4 (EC:2.7.11.1) |
C00044491
|
| 80022 | MYO15B, MYO15BP | myosin XVB pseudogene |
C00044491
|
| 4430 | MYO1B, myr1 | myosin IB |
C00044491
|
| 4641 | MYO1C, MMI-beta, MMIb, NMI, myr2 | myosin IC |
C00044491
|
| 4642 | MYO1D, myr4 | myosin ID |
C00044491
|
| 4643 | MYO1E, FSGS6, HuncM-IC, MYO1C | myosin IE |
C00044491
|
| 4644 | MYO5A, GS1, MYH12, MYO5, MYR12 | myosin VA (heavy chain 12, myoxin) |
C00044491
|
| 4645 | MYO5B | myosin VB |
C00044491
|
| 55930 | MYO5C | myosin VC |
C00044491
|
| 4646 | MYO6, DFNA22, DFNB37 | myosin VI |
C00044491
|
| 745 | MYRF, C11orf9, MRF, Ndt80, pqn-47 | myelin regulatory factor |
C00044491
|
| 25924 | MYRIP, SLAC2-C, SLAC2C | myosin VIIA and Rab interacting protein |
C00044491
|
| 653784 | MZT2A, FAM128A, MOZART2A | mitotic spindle organizing protein 2A |
C00044491
|
| 55728 | N4BP2, B3BP | NEDD4 binding protein 2 |
C00044491
|
| 10443 | N4BP2L2, 92M18.3, CG005, PFAAP5 | NEDD4 binding protein 2-like 2 |
C00044491
|
| 80018 | NAA25, C12orf30, MDM20, NAP1 | N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
C00044491
|
| 64859 | NABP1, OBFC2A, SOSS-B2, SSB2 | nucleic acid binding protein 1 |
C00044491
|
| 138151 | NACC2, BEND9, BTBD14, BTBD14A, BTBD31, RBB | NACC family member 2, BEN and BTB (POZ) domain containing |
C00044491
|
| 65220 | NADK, dJ283E3.1 | NAD kinase (EC:2.7.1.23) |
C00044491
|
| 4668 | NAGA, D22S674, GALB | N-acetylgalactosaminidase, alpha- (EC:3.2.1.49) |
C00044491
|
| 55577 | NAGK, GNK, HSA242910 | N-acetylglucosamine kinase (EC:2.7.1.59) |
C00044491
|
| 342977 | NANOS3, NANOS1L, NOS3 | nanos homolog 3 (Drosophila) |
C00044491
|
| 54187 | NANS, SAS | N-acetylneuraminic acid synthase (EC:2.5.1.56 2.5.1.57) |
C00044491
|
| 4673 | NAP1L1, NAP1, NAP1L, NRP | nucleosome assembly protein 1-like 1 |
C00044491
|
| 222236 | NAPEPLD, FMP30, NAPE-PLD | N-acyl phosphatidylethanolamine phospholipase D (EC:3.1.4.54) |
C00044491
|
| 256236 | NAPSB, NAP1L, NAP2, NAPB, NAPSBP | napsin B aspartic peptidase, pseudogene |
C00044491
|
| 26502 | NARF, IOP2 | nuclear prelamin A recognition factor |
C00044491
|
| 79731 | NARS2, SLM5, asnRS | asparaginyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.22) |
C00044491
|
| 10 | NAT2, AAC2, NAT-2, PNAT | N-acetyltransferase 2 (arylamine N-acetyltransferase) (EC:2.3.1.5) |
C00044491
|
| 9027 | NAT8, ATase2, CML1, GLA, Hcml1, TSC501, TSC510 | N-acetyltransferase 8 (GCN5-related, putative) |
C00044491
|
| 89797 | NAV2, HELAD1, POMFIL2, RAINB1, STEERIN2, UNC53H2 | neuron navigator 2 (EC:3.6.4.12) |
C00044491
|
| 4681 | NBL1, D1S1733E, DAN, DAND1, NB, NO3 | neuroblastoma 1, DAN family BMP antagonist |
C00044491
|
| 4683 | NBN, AT-V1, AT-V2, ATV, NBS, NBS1, P95 | nibrin |
C00044491
|
| 9918 | NCAPD2, CAP-D2, CNAP1, hCAP-D2 | non-SMC condensin I complex, subunit D2 |
C00044491
|
| 23310 | NCAPD3, CAP-D3, CAPD3, hCAP-D3, hHCP-6, hcp-6 | non-SMC condensin II complex, subunit D3 |
C00044491
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00044491
|
| 54892 | NCAPG2, CAP-G2, CAPG2, LUZP5, MTB, hCAP-G2 | non-SMC condensin II complex, subunit G2 |
C00044491
|
| 23397 | NCAPH, BRRN1, CAP-H | non-SMC condensin I complex, subunit H |
C00044491
|
| 29781 | NCAPH2, CAPH2 | non-SMC condensin II complex, subunit H2 |
C00044491
|
| 22916 | NCBP2, CBC2, CBP20, NIP1 | nuclear cap binding protein subunit 2, 20kDa |
C00044491
|
| 342897 | NCCRP1, FBXO50, NCCRP-1 | non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
C00044491
|
| 57552 | NCEH1, AADACL1, NCEH | neutral cholesterol ester hydrolase 1 (EC:3.1.1.-) |
C00044491
|
| 4691 | NCL, C23 | nucleolin |
C00044491
|
| 56926 | NCLN, NET59 | nicalin |
C00044491
|
| 400746 | NCMAP, C1orf130, MP11 | noncompact myelin associated protein |
C00044491
|
| 8648 | NCOA1, F-SRC-1, KAT13A, RIP160, SRC1, bHLHe42, bHLHe74 | nuclear receptor coactivator 1 (EC:2.3.1.48) |
C00044491
|
| 8202 | NCOA3, ACTR, AIB-1, AIB1, CAGH16, CTG26, KAT13B, RAC3, SRC-3, SRC3, TNRC14, TNRC16, TRAM-1, bHLHe42, pCIP | nuclear receptor coactivator 3 (EC:2.3.1.48) |
C00044491
|
| 23054 | NCOA6, AIB3, ASC2, NRC, PRIP, RAP250, TRBP | nuclear receptor coactivator 6 |
C00044491
|
| 135112 | NCOA7, ERAP140, ESNA1, Nbla00052, Nbla10993, TLDC4, dJ187J11.3 | nuclear receptor coactivator 7 |
C00044491
|
| 9611 | NCOR1, N-CoR, N-CoR1, TRAC1, hN-CoR | nuclear receptor corepressor 1 |
C00044491
|
| 9612 | NCOR2, CTG26, N-CoR2, SMAP270, SMRT, SMRTE, SMRTE-tau, TNRC14, TRAC, TRAC-1, TRAC1 | nuclear receptor corepressor 2 |
C00044491
|
| 55706 | NDC1, NET3, TMEM48 | NDC1 transmembrane nucleoporin |
C00044491
|
| 81565 | NDEL1, EOPA, MITAP1, NDE1L1, NDE2, NUDEL | nudE neurodevelopment protein 1-like 1 |
C00044491
|
| 10397 | NDRG1, CAP43, CMT4D, DRG-1, DRG1, GC4, HMSNL, NDR1, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 | N-myc downstream regulated 1 |
C00044491
|
| 3340 | NDST1, HSST, NST1 | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (EC:2.8.2.8) |
C00044491
|
| 4705 | NDUFA10, CI-42KD, CI-42k | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa (EC:1.6.5.3 1.6.99.3) |
C00044491
|
| 4698 | NDUFA5, B13, CI-13KD-B, CI-13kB, NUFM, UQOR13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 (EC:1.6.5.3 1.6.99.3) |
C00044491
|
| 4702 | NDUFA8, CI-19KD, CI-PGIV, PGIV | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa (EC:1.6.5.3 1.6.99.3) |
C00044491
|
| 91942 | NDUFAF2, B17.2L, MMTN, NDUFA12L, mimitin | NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
C00044491
|
| 137682 | NDUFAF6, C8orf38 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
C00044491
|
| 4719 | NDUFS1, CI-75Kd, CI-75k, PRO1304 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) (EC:1.6.5.3 1.6.99.3) |
C00044491
|
| 4726 | NDUFS6, CI-13kA, CI-13kD-A, CI13KDA | NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) (EC:1.6.5.3 1.6.99.3) |
C00044491
|
| 4731 | NDUFV3, CI-10k, CI-9KD | NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa (EC:1.6.5.3) |
C00044491
|
| 10529 | NEBL, LASP2, LNEBL | nebulette |
C00044491
|
| 4734 | NEDD4, NEDD4-1, RPF1 | neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00044491
|
| 23327 | NEDD4L, NEDD4-2, NEDD4.2, RSP5, hNEDD4-2 | neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00044491
|
| 55247 | NEIL3, FGP2, FPG2, NEI3, hFPG2, hNEI3 | nei endonuclease VIII-like 3 (E. coli) (EC:4.2.99.18) |
C00044491
|
| 4750 | NEK1, NY-REN-55, SRPS2, SRPS2A | NIMA-related kinase 1 (EC:2.7.11.1) |
C00044491
|
| 4751 | NEK2, HsPK21, NEK2A, NLK1 | NIMA-related kinase 2 (EC:2.7.11.1) |
C00044491
|
| 4752 | NEK3, HSPK36 | NIMA-related kinase 3 (EC:2.7.11.1) |
C00044491
|
| 6787 | NEK4, NRK2, STK2, pp12301 | NIMA-related kinase 4 (EC:2.7.11.1) |
C00044491
|
| 91754 | NEK9, NERCC, NERCC1, Nek8 | NIMA-related kinase 9 (EC:2.7.11.1) |
C00044491
|
| 4756 | NEO1, IGDCC2, NGN | neogenin 1 |
C00044491
|
| 10276 | NET1, ARHGEF8, NET1A | neuroepithelial cell transforming 1 |
C00044491
|
| 81831 | NETO2, BTCL2, NEOT2 | neuropilin (NRP) and tolloid (TLL)-like 2 |
C00044491
|
| 4758 | NEU1, NANH, NEU, SIAL1 | sialidase 1 (lysosomal sialidase) (EC:3.2.1.18) |
C00044491
|
| 10825 | NEU3, SIAL3 | sialidase 3 (membrane sialidase) (EC:3.2.1.18) |
C00044491
|
| 4762 | NEUROG1, AKA, Math4C, NEUROD3, bHLHa6, ngn1 | neurogenin 1 |
C00044491
|
| 4763 | NF1, NFNS, VRNF, WSS | neurofibromin 1 |
C00044491
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00044491
|
| 4774 | NFIA, CTF, NF-I/A, NF1-A, NFI-A, NFI-L | nuclear factor I/A |
C00044491
|
| 4781 | NFIB, CTF, HMGIC/NFIB, NF-I/B, NF1-B, NFI-B, NFI-RED, NFIB2, NFIB3 | nuclear factor I/B |
C00044491
|
| 4782 | NFIC, CTF, CTF5, NF-I, NFI | nuclear factor I/C (CCAAT-binding transcription factor) |
C00044491
|
| 4783 | NFIL3, E4BP4, IL3BP1, NF-IL3A, NFIL3A | nuclear factor, interleukin 3 regulated |
C00044491
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00044491
|
| 4795 | NFKBIL1, IKBL, LST1, NFKBIL | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
C00044491
|
| 64332 | NFKBIZ, IKBZ, INAP, MAIL | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
C00044491
|
| 4799 | NFX1, NFX2 | nuclear transcription factor, X-box binding 1 (EC:6.3.2.-) |
C00044491
|
| 4800 | NFYA, CBF-A, CBF-B, HAP2, NF-YA | nuclear transcription factor Y, alpha |
C00044491
|
| 4801 | NFYB, CBF-A, CBF-B, HAP3, NF-YB | nuclear transcription factor Y, beta |
C00044491
|
| 25791 | NGEF, ARHGEF27, EPHEXIN | neuronal guanine nucleotide exchange factor |
C00044491
|
| 55768 | NGLY1, CDG1V, PNG1, PNGase | N-glycanase 1 (EC:3.5.1.52) |
C00044491
|
| 387921 | NHLRC3 | NHL repeat containing 3 |
C00044491
|
| 57224 | NHSL1, C6orf63 | NHS-like 1 |
C00044491
|
| 4811 | NID1, NID | nidogen 1 |
C00044491
|
| 60491 | NIF3L1, ALS2CR1, CALS-7 | NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
C00044491
|
| 51199 | NIN, SCKL7 | ninein (GSK3B interacting protein) |
C00044491
|
| 4815 | NINJ2 | ninjurin 2 |
C00044491
|
| 79815 | NIPAL2, NPAL2 | NIPA-like domain containing 2 |
C00044491
|
| 390010 | NKX1-2, C10orf121, NKX-1.1, bB238F13.2 | NK1 homeobox 2 |
C00044491
|
| 7080 | NKX2-1, BCH, BHC, NK-2, NKX2.1, NKX2A, TEBP, TITF1, TTF-1, TTF1 | NK2 homeobox 1 |
C00044491
|
| 4824 | NKX3-1, BAPX2, NKX3, NKX3.1, NKX3A | NK3 homeobox 1 |
C00044491
|
| 51701 | NLK | nemo-like kinase (EC:2.7.11.24) |
C00044491
|
| 57486 | NLN, AGTBP, EP24.16, MEP, MOP | neurolysin (metallopeptidase M3 family) (EC:3.4.24.16) |
C00044491
|
| 84166 | NLRC5, CLR16.1, NOD27, NOD4 | NLR family, CARD domain containing 5 |
C00044491
|
| 126204 | NLRP13, CLR19.7, NALP13, NOD14, PAN13 | NLR family, pyrin domain containing 13 |
C00044491
|
| 283458 |
C00044491
|
||
| 4833 | NME4, NDPK-D, NM23H4, nm23-H4 | NME/NM23 nucleoside diphosphate kinase 4 (EC:2.7.4.6) |
C00044491
|
| 10201 | NME6, IPIA-ALPHA, NDK_6, NM23-H6 | NME/NM23 nucleoside diphosphate kinase 6 (EC:2.7.4.6) |
C00044491
|
| 29922 | NME7, MN23H7, NDK_7, NDK7, nm23-H7 | NME/NM23 family member 7 (EC:2.7.4.6) |
C00044491
|
| 9111 | NMI | N-myc (and STAT) interactor |
C00044491
|
| 54981 | NMRK1, C9orf95, NRK1, bA235O14.2 | nicotinamide riboside kinase 1 (EC:2.7.1.22 2.7.1.173) |
C00044491
|
| 4836 | NMT1, NMT | N-myristoyltransferase 1 (EC:2.3.1.97) |
C00044491
|
| 4837 | NNMT | nicotinamide N-methyltransferase (EC:2.1.1.1) |
C00044491
|
| 10392 | NOD1, CARD4, CLR7.1, NLRC1 | nucleotide-binding oligomerization domain containing 1 |
C00044491
|
| 79954 | NOL10, PQBP5 | nucleolar protein 10 |
C00044491
|
| 8996 | NOL3, ARC, FCM, MYP, NOP, NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) |
C00044491
|
| 55035 | NOL8, C9orf34, NOP132, bA62C3.3, bA62C3.4 | nucleolar protein 8 |
C00044491
|
| 79707 | NOL9, Grc3, NET6 | nucleolar protein 9 |
C00044491
|
| 64434 | NOM1, C7orf3, SGD1 | nucleolar protein with MIF4G domain 1 |
C00044491
|
| 51491 | NOP16, HSPC111, HSPC185 | NOP16 nucleolar protein |
C00044491
|
| 51070 | NOSIP | nitric oxide synthase interacting protein |
C00044491
|
| 4851 | NOTCH1, TAN1, hN1 | notch 1 |
C00044491
|
| 4853 | NOTCH2, AGS2, HJCYS, hN2 | notch 2 |
C00044491
|
| 4855 | NOTCH4, INT3, NOTCH3 | notch 4 |
C00044491
|
| 4857 | NOVA1, Nova-1 | neuro-oncological ventral antigen 1 |
C00044491
|
| 4863 | NPAT, E14, E14/NPAT, p220 | nuclear protein, ataxia-telangiectasia locus |
C00044491
|
| 256933 | NPB, L7, PPL7, PPNPB | neuropeptide B |
C00044491
|
| 4864 | NPC1, NPC | Niemann-Pick disease, type C1 |
C00044491
|
| 29881 | NPC1L1, NPC11L1 | NPC1-like 1 |
C00044491
|
| 55666 | NPLOC4, NPL4 | nuclear protein localization 4 homolog (S. cerevisiae) |
C00044491
|
| 255743 | NPNT, EGFL6L, POEM | nephronectin |
C00044491
|
| 4887 | NPY2R, NPY2-R | neuropeptide Y receptor Y2 |
C00044491
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00044491
|
| 9975 | NR1D2, BD73, EAR-1R, RVR | nuclear receptor subfamily 1, group D, member 2 |
C00044491
|
| 9971 | NR1H4, BAR, FXR, HRR-1, HRR1, RIP14 | nuclear receptor subfamily 1, group H, member 4 |
C00044491
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00044491
|
| 9970 | NR1I3, CAR, CAR1, MB67 | nuclear receptor subfamily 1, group I, member 3 |
C00044491
|
| 7181 | NR2C1, TR2 | nuclear receptor subfamily 2, group C, member 1 |
C00044491
|
| 7025 | NR2F1, COUP-TFI, EAR-3, EAR3, ERBAL3, NR2F2, SVP44, TCFCOUP1, TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 |
C00044491
|
| 7026 | NR2F2, ARP1, COUPTFB, COUPTFII, NF-E3, NR2F1, SVP40, TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 |
C00044491
|
| 2063 | NR2F6, EAR-2, EAR2, ERBAL2 | nuclear receptor subfamily 2, group F, member 6 |
C00044491
|
| 2908 | NR3C1, GCCR, GCR, GR, GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
C00044491
|
| 2494 | NR5A2, B1F, B1F2, CPF, FTF, FTZ-F1, FTZ-F1beta, LRH-1, LRH1, hB1F-2 | nuclear receptor subfamily 5, group A, member 2 |
C00044491
|
| 4893 | NRAS, ALPS4, N-ras, NRAS1, NS6 | neuroblastoma RAS viral (v-ras) oncogene homolog |
C00044491
|
| 4898 | NRD1, hNRD1, hNRD2 | nardilysin (N-arginine dibasic convertase) (EC:3.4.24.61) |
C00044491
|
| 9315 | NREP, C5orf13, D4S114, P311, PRO1873, PTZ17, SEZ17 | neuronal regeneration related protein |
C00044491
|
| 9542 | NRG2, DON1, HRG2, NTAK | neuregulin 2 |
C00044491
|
| 8829 | NRP1, BDCA4, CD304, NP1, NRP, VEGF165R | neuropilin 1 |
C00044491
|
| 140767 | NRSN1, VMP, p24 | neurensin 1 |
C00044491
|
| 9379 | NRXN2 | neurexin 2 |
C00044491
|
| 4905 | NSF, SKD2 | N-ethylmaleimide-sensitive factor (EC:3.6.4.6) |
C00044491
|
| 54780 | NSMCE4A, C10orf86, NS4EA, NSE4A | non-SMC element 4 homolog A (S. cerevisiae) |
C00044491
|
| 55695 | NSUN5, NOL1, NOL1R, NSUN5A, WBSCR20, WBSCR20A, p120 | NOP2/Sun domain family, member 5 |
C00044491
|
| 221078 | NSUN6, 4933414E04Rik, NOPD1 | NOP2/Sun domain family, member 6 |
C00044491
|
| 30833 | NT5C, DNT, DNT1, P5N2, PN-I, PN-II, UMPH2, cdN, dNT-1 | 5', 3'-nucleotidase, cytosolic (EC:3.1.3.5) |
C00044491
|
| 64943 | NT5DC2 | 5'-nucleotidase domain containing 2 (EC:3.1.3.5) |
C00044491
|
| 4907 | NT5E, CALJA, CD73, E5NT, NT, NT5, NTE, eN, eNT | 5'-nucleotidase, ecto (CD73) (EC:3.1.3.5) |
C00044491
|
| 4908 | NTF3, HDNF, NGF-2, NGF2, NT3 | neurotrophin 3 |
C00044491
|
| 50863 | NTM, HNT, IGLON2, NTRI | neurotrimin |
C00044491
|
| 59277 | NTN4, PRO3091 | netrin 4 |
C00044491
|
| 84628 | NTNG2, LHLL9381, Lmnt2, NTNG1, bA479K20.1 | netrin G2 |
C00044491
|
| 84284 | NTPCR, C1orf57, HCR-NTPase, RP4-678E16.2 | nucleoside-triphosphatase, cancer-related (EC:3.6.1.15) |
C00044491
|
| 51667 | NUB1, BS4, NUB1L, NYREN18 | negative regulator of ubiquitin-like proteins 1 |
C00044491
|
| 4925 | NUCB2, NEFA | nucleobindin 2 |
C00044491
|
| 4521 | NUDT1, MTH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 (EC:3.6.1.55 3.6.1.56) |
C00044491
|
| 83594 | NUDT12 | nudix (nucleoside diphosphate linked moiety X)-type motif 12 (EC:3.6.1.22) |
C00044491
|
| 11051 | NUDT21, CFIM25, CPSF5 | nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
C00044491
|
| 11164 | NUDT5, YSA1, YSA1H, YSAH1, hYSAH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 5 (EC:3.6.1.13 3.6.1.58) |
C00044491
|
| 283927 | NUDT7 | nudix (nucleoside diphosphate linked moiety X)-type motif 7 (EC:3.6.1.-) |
C00044491
|
| 254552 | NUDT8 | nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
C00044491
|
| 53343 | NUDT9, NUDT10 | nudix (nucleoside diphosphate linked moiety X)-type motif 9 (EC:3.6.1.13) |
C00044491
|
| 83540 | NUF2, CDCA1, CT106, NUF2R | NUF2, NDC80 kinetochore complex component |
C00044491
|
| 57532 | NUFIP2, 182-FIP, 82-FIP, FIP-82 | nuclear fragile X mental retardation protein interacting protein 2 |
C00044491
|
| 4926 | NUMA1, NUMA | nuclear mitotic apparatus protein 1 |
C00044491
|
| 57122 | NUP107, NUP84 | nucleoporin 107kDa |
C00044491
|
| 55746 | NUP133, hNUP133 | nucleoporin 133kDa |
C00044491
|
| 9972 | NUP153, HNUP153, N153 | nucleoporin 153kDa |
C00044491
|
| 23511 | NUP188, KIAA0169, hNup188 | nucleoporin 188kDa |
C00044491
|
| 23165 | NUP205, C7orf14 | nucleoporin 205kDa |
C00044491
|
| 23225 | NUP210, GP210, POM210 | nucleoporin 210kDa |
C00044491
|
| 8021 | NUP214, CAIN, CAN, D9S46E, N214, p250 | nucleoporin 214kDa |
C00044491
|
| 129401 | NUP35, MP44, NP44, NUP53 | nucleoporin 35kDa |
C00044491
|
| 79023 | NUP37, p37 | nucleoporin 37kDa |
C00044491
|
| 10762 | NUP50, NPAP60, NPAP60L | nucleoporin 50kDa |
C00044491
|
| 23636 | NUP62, IBSN, SNDI, p62 | nucleoporin 62kDa |
C00044491
|
| 54830 | NUP62CL | nucleoporin 62kDa C-terminal like |
C00044491
|
| 79902 | NUP85, Nup75 | nucleoporin 85kDa |
C00044491
|
| 4927 | NUP88 | nucleoporin 88kDa |
C00044491
|
| 4928 | NUP98, ADIR2, NUP196, NUP96 | nucleoporin 98kDa |
C00044491
|
| 9818 | NUPL1, PRO2463 | nucleoporin like 1 |
C00044491
|
| 51203 | NUSAP1, ANKT, BM037, LNP, NUSAP, PRO0310p1, Q0310, SAPL | nucleolar and spindle associated protein 1 |
C00044491
|
| 10482 | NXF1, MEX67, TAP | nuclear RNA export factor 1 |
C00044491
|
| 158046 | NXNL2, C9orf121, RDCVF2 | nucleoredoxin-like 2 (EC:1.8.1.8) |
C00044491
|
| 54827 | NXPE4, C11orf33, FAM55D | neurexophilin and PC-esterase domain family, member 4 |
C00044491
|
| 11248 | NXPH3, NPH3 | neurexophilin 3 |
C00044491
|
| 220323 | OAF, NS5ATP13TP2 | OAF homolog (Drosophila) |
C00044491
|
| 221443 | OARD1, C6orf130, TARG1, dJ34B21.3 | O-acyl-ADP-ribose deacylase 1 |
C00044491
|
| 4938 | OAS1, IFI-4, OIAS, OIASI | 2'-5'-oligoadenylate synthetase 1, 40/46kDa (EC:2.7.7.84) |
C00044491
|
| 4940 | OAS3, p100 | 2'-5'-oligoadenylate synthetase 3, 100kDa (EC:2.7.7.84) |
C00044491
|
| 4942 | OAT, GACR, HOGA, OATASE, OKT | ornithine aminotransferase (EC:2.6.1.13) |
C00044491
|
| 23363 | OBSL1 | obscurin-like 1 (EC:2.7.11.18) |
C00044491
|
| 132299 | OCIAD2 | OCIA domain containing 2 |
C00044491
|
| 54959 | ODAM, APIN | odontogenic, ameloblast asssociated |
C00044491
|
| 4957 | ODF2, CT134, ODF2/1, ODF2/2, ODF84 | outer dense fiber of sperm tails 2 |
C00044491
|
| 8473 | OGT, HRNT1, O-GLCNAC | O-linked N-acetylglucosamine (GlcNAc) transferase (EC:2.4.1.255) |
C00044491
|
| 11339 | OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta, hMIS18beta | Opa interacting protein 5 |
C00044491
|
| 729082 | OIP5-AS1, cyrano | OIP5 antisense RNA 1 |
C00044491
|
| 170392 | OIT3, LZP | oncoprotein induced transcript 3 |
C00044491
|
| 116448 | OLIG1, BHLHB6, BHLHE21 | oligodendrocyte transcription factor 1 |
C00044491
|
| 115209 | OMA1, 2010001O09Rik, DAB1, MPRP-1, YKR087C, ZMPOMA1, peptidase | OMA1 zinc metallopeptidase |
C00044491
|
| 9480 | ONECUT2, OC-2, OC2 | one cut homeobox 2 |
C00044491
|
| 4976 | OPA1, MGM1, NPG, NTG, largeG | optic atrophy 1 (autosomal dominant) (EC:3.6.5.5) |
C00044491
|
| 10133 | OPTN, ALS12, FIP2, GLC1E, HIP7, HYPL, NRP, TFIIIA-INTP | optineurin |
C00044491
|
| 390264 | OR10G4, OR11-278 | olfactory receptor, family 10, subfamily G, member 4 |
C00044491
|
| 81697 | OR2B2, OR2B2Q, OR2B9, OR6-1, dJ193B12.4, hs6M1-10 | olfactory receptor, family 2, subfamily B, member 2 |
C00044491
|
| 391211 | OR2G6 | olfactory receptor, family 2, subfamily G, member 6 |
C00044491
|
| 7932 | OR2H2, FAT11, OLFR2, OLFR42B, OR2H3, dJ271M21.2, hs6M1-12 | olfactory receptor, family 2, subfamily H, member 2 |
C00044491
|
| 442186 | OR2J3, 6M1-3, C3HEXS, HS6M1-3, OR6-16, OR6-6, OR6.3.6, ORL671 | olfactory receptor, family 2, subfamily J, member 3 |
C00044491
|
| 26246 | OR2L2, HSHTPCRH07, HTPCRH07, OR1-48, OR2L12, OR2L4P | olfactory receptor, family 2, subfamily L, member 2 |
C00044491
|
| 391196 | OR2M7, OR1-58 | olfactory receptor, family 2, subfamily M, member 7 |
C00044491
|
| 26692 | OR2W1, hs6M1-15 | olfactory receptor, family 2, subfamily W, member 1 |
C00044491
|
| 26689 | OR4D1, OR17-23, OR4D3, OR4D4P, TPCR16 | olfactory receptor, family 4, subfamily D, member 1 |
C00044491
|
| 390072 | OR52N4, OR11-64 | olfactory receptor, family 52, subfamily N, member 4 |
C00044491
|
| 23595 | ORC3, LAT, LATHEO, ORC3L | origin recognition complex, subunit 3 |
C00044491
|
| 5001 | ORC5, ORC5L, ORC5P, ORC5T | origin recognition complex, subunit 5 |
C00044491
|
| 716175 |
C00044491
|
||
| 94103 | ORMDL3 | ORM1-like 3 (S. cerevisiae) |
C00044491
|
| 23762 | OSBP2, HLM, ORP-4, ORP4, OSBPL1, OSBPL4 | oxysterol binding protein 2 |
C00044491
|
| 114884 | OSBPL10, ORP10, OSBP9 | oxysterol binding protein-like 10 |
C00044491
|
| 26031 | OSBPL3, ORP-3, ORP3, OSBP3 | oxysterol binding protein-like 3 |
C00044491
|
| 114880 | OSBPL6, ORP6 | oxysterol binding protein-like 6 |
C00044491
|
| 114881 | OSBPL7, ORP7 | oxysterol binding protein-like 7 |
C00044491
|
| 51526 | OSER1, C20orf111, HSPC207, Osr1, Perit1, dJ1183I21.1 | oxidative stress responsive serine-rich 1 |
C00044491
|
| 29948 | OSGIN1, BDGI, OKL38 | oxidative stress induced growth inhibitor 1 |
C00044491
|
| 9180 | OSMR, OSMRB, PLCA1 | oncostatin M receptor |
C00044491
|
| 58505 | OSTC | oligosaccharyltransferase complex subunit (non-catalytic) |
C00044491
|
| 28962 | OSTM1, GIPN, GL, OPTB5 | osteopetrosis associated transmembrane protein 1 |
C00044491
|
| 92736 | OTOP2 | otopetrin 2 |
C00044491
|
| 220213 | OTUD1, DUBA7, OTDC1 | OTU domain containing 1 (EC:3.4.19.12) |
C00044491
|
| 23252 | OTUD3, DUBA4, RP11-460G22.1 | OTU domain containing 3 (EC:3.4.19.12) |
C00044491
|
| 54726 | OTUD4, DUBA6, HIN1, HSHIN1 | OTU domain containing 4 (EC:3.4.19.12) |
C00044491
|
| 161725 | OTUD7A, C15orf16, C16ORF15, CEZANNE2, OTUD7 | OTU domain containing 7A (EC:3.4.19.12) |
C00044491
|
| 55074 | OXR1, TLDC3 | oxidation resistance 1 |
C00044491
|
| 5025 | P2RX4, P2X4, P2X4R | purinergic receptor P2X, ligand-gated ion channel, 4 |
C00044491
|
| 8974 | P4HA2 | prolyl 4-hydroxylase, alpha polypeptide II (EC:1.14.11.2) |
C00044491
|
| 23241 | PACS2, PACS-2, PACS1L | phosphofurin acidic cluster sorting protein 2 |
C00044491
|
| 29993 | PACSIN1, SDPI | protein kinase C and casein kinase substrate in neurons 1 |
C00044491
|
| 11252 | PACSIN2, SDPII | protein kinase C and casein kinase substrate in neurons 2 |
C00044491
|
| 5048 | PAFAH1B1, LIS1, LIS2, MDCR, MDS, PAFAH | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) (EC:3.1.1.47) |
C00044491
|
| 5051 | PAFAH2, HSD-PLA2 | platelet-activating factor acetylhydrolase 2, 40kDa (EC:3.1.1.47) |
C00044491
|
| 55824 | PAG1, CBP, PAG | phosphoprotein associated with glycosphingolipid microdomains 1 |
C00044491
|
| 203569 | PAGE2, CT16.4, GAGEC2, GAGEE2, PAGE-2 | P antigen family, member 2 (prostate associated) |
C00044491
|
| 139793 | PAGE3, CT16.6, GAGED1, PAGE-3 | P antigen family, member 3 (prostate associated) |
C00044491
|
| 10605 | PAIP1 | poly(A) binding protein interacting protein 1 |
C00044491
|
| 57144 | PAK7, PAK5 | p21 protein (Cdc42/Rac)-activated kinase 7 (EC:2.7.11.1) |
C00044491
|
| 5064 | PALM | paralemmin |
C00044491
|
| 114299 | PALM2, AKAP2 | paralemmin 2 |
C00044491
|
| 54873 | PALMD, C1orf11, PALML | palmdelphin |
C00044491
|
| 9924 | PAN2, USP52 | PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (EC:3.1.13.4) |
C00044491
|
| 24145 | PANX1, MRS1, PX1, UNQ2529 | pannexin 1 |
C00044491
|
| 56666 | PANX2, PX2, hPANX2 | pannexin 2 |
C00044491
|
| 167153 | PAPD4, GLD2 | PAP associated domain containing 4 (EC:2.7.7.19) |
C00044491
|
| 64282 | PAPD5, TRF4-2 | PAP associated domain containing 5 |
C00044491
|
| 11044 | PAPD7, LAK-1, LAK1, POLK, POLS, TRF4, TRF4-1, TRF41, TUTASE5 | PAP associated domain containing 7 (EC:2.7.7.7) |
C00044491
|
| 493913 | PAPPA-AS1, DIPAS, NCRNA00156, PAPPA-AS, PAPPAAS, PAPPAS, TAF2C2, TAF4B, TAFII105 | PAPPA antisense RNA 1 |
C00044491
|
| 9060 | PAPSS2, ATPSK2, BCYM4, SK2 | 3'-phosphoadenosine 5'-phosphosulfate synthase 2 (EC:2.7.1.25 2.7.7.4) |
C00044491
|
| 79957 | PAQR6 | progestin and adipoQ receptor family member VI |
C00044491
|
| 344838 | PAQR9 | progestin and adipoQ receptor family member IX |
C00044491
|
| 84552 | PARD6G, PAR-6G, PAR6gamma | par-6 family cell polarity regulator gamma |
C00044491
|
| 100130522 | PARD6G-AS1 | PARD6G antisense RNA 1 |
C00044491
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00044491
|
| 64761 | PARP12, ARTD12, MST109, MSTP109, ZC3H1, ZC3HDC1 | poly (ADP-ribose) polymerase family, member 12 (EC:2.4.2.30) |
C00044491
|
| 54625 | PARP14, ARTD8, BAL2, PARP-14, pART8 | poly (ADP-ribose) polymerase family, member 14 (EC:2.4.2.30) |
C00044491
|
| 10038 | PARP2, ADPRT2, ADPRTL2, ADPRTL3, ARTD2, PARP-2, pADPRT-2 | poly (ADP-ribose) polymerase 2 (EC:2.4.2.30) |
C00044491
|
| 143 | PARP4, ADPRTL1, ARTD4, PARP-4, PARPL, PH5P, VAULT3, VPARP, VWA5C, p193 | poly (ADP-ribose) polymerase family, member 4 (EC:2.4.2.30) |
C00044491
|
| 83666 | PARP9, ARTD9, BAL, BAL1, MGC:7868 | poly (ADP-ribose) polymerase family, member 9 (EC:2.4.2.30) |
C00044491
|
| 55010 | PARPBP, AROM, C12orf48, PARI | PARP1 binding protein |
C00044491
|
| 23178 | PASK, PASKIN, STK37 | PAS domain containing serine/threonine kinase (EC:2.7.11.1) |
C00044491
|
| 219988 | PATL1, Pat1b, hPat1b | protein associated with topoisomerase II homolog 1 (yeast) |
C00044491
|
| 5075 | PAX1, HUP48 | paired box 1 |
C00044491
|
| 94104 | PAXBP1, BM020, C21orf66, FSAP105, GCFC, GCFC1 | PAX3 and PAX7 binding protein 1 |
C00044491
|
| 51260 | PBDC1, CXorf26 | polysaccharide biosynthesis domain containing 1 |
C00044491
|
| 5089 | PBX2, G17, HOX12, PBX2MHC | pre-B-cell leukemia homeobox 2 |
C00044491
|
| 5090 | PBX3 | pre-B-cell leukemia homeobox 3 |
C00044491
|
| 5094 | PCBP2, HNRNPE2, HNRPE2, hnRNP-E2 | poly(rC) binding protein 2 |
C00044491
|
| 5097 | PCDH1, PC42, PCDH42 | protocadherin 1 |
C00044491
|
| 64773 | PCED1A, C20orf81, FAM113A, bA12M19.1 | PC-esterase domain containing 1A |
C00044491
|
| 7703 | PCGF2, MEL-18, RNF110, ZNF144 | polycomb group ring finger 2 |
C00044491
|
| 55795 | PCID2, F10 | PCI domain containing 2 |
C00044491
|
| 5105 | PCK1, PEPCK-C, PEPCK1, PEPCKC | phosphoenolpyruvate carboxykinase 1 (soluble) (EC:4.1.1.32) |
C00044491
|
| 5108 | PCM1, PTC4 | pericentriolar material 1 |
C00044491
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00044491
|
| 57092 | PCNP | PEST proteolytic signal containing nuclear protein |
C00044491
|
| 22990 | PCNX, PCNXL1 | pecanex homolog (Drosophila) |
C00044491
|
| 26577 | PCOLCE2, PCPE2 | procollagen C-endopeptidase enhancer 2 |
C00044491
|
| 126006 | PCP2, GPSM4 | Purkinje cell protein 2 |
C00044491
|
| 255738 | PCSK9, FH3, HCHOLA3, LDLCQ1, NARC-1, NARC1, PC9 | proprotein convertase subtilisin/kexin type 9 |
C00044491
|
| 27250 | PDCD4, H731 | programmed cell death 4 (neoplastic transformation inhibitor) |
C00044491
|
| 10015 | PDCD6IP, AIP1, ALIX, DRIP4, HP95 | programmed cell death 6 interacting protein |
C00044491
|
| 5082 | PDCL, PhLP | phosducin-like |
C00044491
|
| 5143 | PDE4C, DPDE1 | phosphodiesterase 4C, cAMP-specific (EC:3.1.4.17) |
C00044491
|
| 9659 | PDE4DIP, CMYA2, MMGL | phosphodiesterase 4D interacting protein |
C00044491
|
| 5154 | PDGFA, PDGF-A, PDGF1 | platelet-derived growth factor alpha polypeptide |
C00044491
|
| 10954 | PDIA5, PDIR | protein disulfide isomerase family A, member 5 (EC:5.3.4.1) |
C00044491
|
| 5165 | PDK3, CMTX6 | pyruvate dehydrogenase kinase, isozyme 3 (EC:2.7.11.2) |
C00044491
|
| 9124 | PDLIM1, CLIM1, CLP-36, CLP36, hCLIM1 | PDZ and LIM domain 1 |
C00044491
|
| 10611 | PDLIM5, ENH, ENH1, LIM | PDZ and LIM domain 5 |
C00044491
|
| 5170 | PDPK1, PDK1, PDPK2, PRO0461 | 3-phosphoinositide dependent protein kinase-1 (EC:2.7.11.1) |
C00044491
|
| 23244 | PDS5A, SCC-112, SCC112 | PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
C00044491
|
| 23047 | PDS5B, APRIN, AS3, CG008 | PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
C00044491
|
| 23590 | PDSS1, COQ1, COQ10D2, DPS, RP13-16H11.3, SPS, TPRT, TPT, TPT_1, hDPS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 (EC:2.5.1.91) |
C00044491
|
| 23042 | PDXDC1, LP8165 | pyridoxal-dependent decarboxylase domain containing 1 |
C00044491
|
| 8566 | PDXK, C21orf124, C21orf97, PKH, PNK | pyridoxal (pyridoxine, vitamin B6) kinase (EC:2.7.1.35) |
C00044491
|
| 5174 | PDZK1, CAP70, CLAMP, NHERF-3, NHERF3, PDZD1 | PDZ domain containing 1 |
C00044491
|
| 8682 | PEA15, HMAT1, HUMMAT1H, MAT1, MAT1H, PEA-15, PED | phosphoprotein enriched in astrocytes 15 |
C00044491
|
| 5175 | PECAM1, CD31, CD31/EndoCAM, GPIIA', PECA1, PECAM-1, endoCAM | platelet/endothelial cell adhesion molecule 1 |
C00044491
|
| 23089 | PEG10, EDR, HB-1, MEF3L, Mar2, Mart2, RGAG3 | paternally expressed 10 |
C00044491
|
| 10400 | PEMT, PEAMT, PEMPT, PEMT2, PNMT | phosphatidylethanolamine N-methyltransferase (EC:2.1.1.17 2.1.1.71) |
C00044491
|
| 5184 | PEPD, PROLIDASE | peptidase D (EC:3.4.13.9) |
C00044491
|
| 5187 | PER1, PER, RIGUI, hPER | period circadian clock 1 |
C00044491
|
| 8864 | PER2, FASPS, FASPS1 | period circadian clock 2 |
C00044491
|
| 8863 | PER3, GIG13 | period circadian clock 3 |
C00044491
|
| 5189 | PEX1, PBD1A, PBD1B, ZWS, ZWS1 | peroxisomal biogenesis factor 1 |
C00044491
|
| 8799 | PEX11B, PEX11-BETA, PEX14B | peroxisomal biogenesis factor 11 beta |
C00044491
|
| 5193 | PEX12, PAF-3, PBD3A | peroxisomal biogenesis factor 12 |
C00044491
|
| 5194 | PEX13, NALD, PBD11A, PBD11B, ZWS | peroxisomal biogenesis factor 13 |
C00044491
|
| 8504 | PEX3, PBD10A, TRG18 | peroxisomal biogenesis factor 3 |
C00044491
|
| 5198 | PFAS, FGAMS, FGARAT, PURL | phosphoribosylformylglycinamidine synthase (EC:6.3.5.3) |
C00044491
|
| 5207 | PFKFB1, F6PK, HL2K, PFRX | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (EC:2.7.1.105 3.1.3.46) |
C00044491
|
| 5213 | PFKM, GSD7, PFK-1, PFK1, PFKA, PFKX | phosphofructokinase, muscle (EC:2.7.1.11) |
C00044491
|
| 80055 | PGAP1, Bst1, ISPD3024 | post-GPI attachment to proteins 1 |
C00044491
|
| 84547 | PGBD1, HUCEP-4, SCAND4, dJ874C20.4 | piggyBac transposable element derived 1 |
C00044491
|
| 5226 | PGD, 6PGD | phosphogluconate dehydrogenase (EC:1.1.1.44) |
C00044491
|
| 5228 | PGF, D12S1900, PGFL, PLGF, PlGF-2, SHGC-10760 | placental growth factor |
C00044491
|
| 5230 | PGK1, MIG10, PGKA | phosphoglycerate kinase 1 (EC:2.7.2.3) |
C00044491
|
| 5236 | PGM1, CDG1T, GSD14 | phosphoglucomutase 1 (EC:5.4.2.2) |
C00044491
|
| 55276 | PGM2 | phosphoglucomutase 2 (EC:5.4.2.2 5.4.2.7) |
C00044491
|
| 283209 | PGM2L1, BM32A | phosphoglucomutase 2-like 1 (EC:2.7.1.106) |
C00044491
|
| 54858 | PGPEP1, PGP, PGP-I, PGPI, Pcp | pyroglutamyl-peptidase I (EC:3.4.19.3) |
C00044491
|
| 10857 | PGRMC1, HPR6.6, MPR | progesterone receptor membrane component 1 |
C00044491
|
| 221692 | PHACTR1, RPEL, RPEL1, dJ257A7.2 | phosphatase and actin regulator 1 |
C00044491
|
| 9749 | PHACTR2, C6orf56 | phosphatase and actin regulator 2 |
C00044491
|
| 1911 | PHC1, EDR1, HPH1, MCPH11, RAE28 | polyhomeotic homolog 1 (Drosophila) |
C00044491
|
| 80012 | PHC3, EDR3, HPH3 | polyhomeotic homolog 3 (Drosophila) |
C00044491
|
| 51131 | PHF11, APY, BCAP, IGEL, IGER, IGHER, NY-REN-34, NYREN34 | PHD finger protein 11 |
C00044491
|
| 148479 | PHF13, PHF5, SPOC1 | PHD finger protein 13 |
C00044491
|
| 9678 | PHF14 | PHD finger protein 14 |
C00044491
|
| 51230 | PHF20, C20orf104, GLEA2, HCA58, NZF, TDRD20A, TZP | PHD finger protein 20 |
C00044491
|
| 112885 | PHF21B, BHC80L, PHF4 | PHD finger protein 21B |
C00044491
|
| 79142 | PHF23, hJUNE-1b | PHD finger protein 23 |
C00044491
|
| 84295 | PHF6, BFLS, BORJ, CENP-31 | PHD finger protein 6 |
C00044491
|
| 51533 | PHF7, HSPC045, HSPC226, NYD-SP6 | PHD finger protein 7 |
C00044491
|
| 5256 | PHKA2, GSD9A, PHK, PYK, PYKL, XLG, XLG2 | phosphorylase kinase, alpha 2 (liver) (EC:2.7.11.19) |
C00044491
|
| 5257 | PHKB | phosphorylase kinase, beta (EC:2.7.11.19) |
C00044491
|
| 90102 | PHLDB2, LL5b, LL5beta | pleckstrin homology-like domain, family B, member 2 |
C00044491
|
| 23035 | PHLPP2, PHLPPL | PH domain and leucine rich repeat protein phosphatase 2 (EC:3.1.3.16) |
C00044491
|
| 57157 | PHTF2 | putative homeodomain transcription factor 2 |
C00044491
|
| 5264 | PHYH, LN1, LNAP1, PAHX, PHYH1, RD | phytanoyl-CoA 2-hydroxylase (EC:1.14.11.18) |
C00044491
|
| 85007 | PHYKPL, AGXT2L2, PHLU | 5-phosphohydroxy-L-lysine phospho-lyase (EC:4.2.3.134) |
C00044491
|
| 51050 | PI15, CRISP8, P24TI, P25TI | peptidase inhibitor 15 |
C00044491
|
| 221476 | PI16, CRISP9, MSMBBP, PSPBP | peptidase inhibitor 16 |
C00044491
|
| 55361 | PI4K2A, PI4KII, PIK42A, RP11-548K23.6 | phosphatidylinositol 4-kinase type 2 alpha (EC:2.7.1.67) |
C00044491
|
| 55300 | PI4K2B, PI4KIIB, PIK42B | phosphatidylinositol 4-kinase type 2 beta (EC:2.7.1.67) |
C00044491
|
| 5298 | PI4KB, NPIK, PI4K-BETA, PI4K92, PI4KBETA, PI4KIIIBETA, PIK4CB | phosphatidylinositol 4-kinase, catalytic, beta (EC:2.7.1.67) |
C00044491
|
| 8554 | PIAS1, DDXBP1, GBP, GU/RH-II, ZMIZ3 | protein inhibitor of activated STAT, 1 |
C00044491
|
| 10464 | PIBF1, C13orf24, CEP90, PIBF, RP11-505F3.1 | progesterone immunomodulatory binding factor 1 |
C00044491
|
| 9780 | PIEZO1, DHS, FAM38A, Mib | piezo-type mechanosensitive ion channel component 1 |
C00044491
|
| 5277 | PIGA, GPI3, MCAHS2, PIG-A | phosphatidylinositol glycan anchor biosynthesis, class A (EC:2.4.1.198) |
C00044491
|
| 5279 | PIGC, GPI2 | phosphatidylinositol glycan anchor biosynthesis, class C (EC:2.4.1.198) |
C00044491
|
| 10026 | PIGK, GPI8 | phosphatidylinositol glycan anchor biosynthesis, class K (EC:3.-.-.-) |
C00044491
|
| 93183 | PIGM, GPI-MT-I | phosphatidylinositol glycan anchor biosynthesis, class M (EC:2.4.1.-) |
C00044491
|
| 51604 | PIGT, MCAHS3, NDAP, PNH2 | phosphatidylinositol glycan anchor biosynthesis, class T |
C00044491
|
| 284098 | PIGW, Gwt1 | phosphatidylinositol glycan anchor biosynthesis, class W |
C00044491
|
| 54965 | PIGX, PIG-X | phosphatidylinositol glycan anchor biosynthesis, class X |
C00044491
|
| 80235 | PIGZ, GPI-MT-IV, PIG-Z, SMP3 | phosphatidylinositol glycan anchor biosynthesis, class Z |
C00044491
|
| 5286 | PIK3C2A, CPK, PI3-K-C2(ALPHA), PI3-K-C2A | phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha (EC:2.7.1.154) |
C00044491
|
| 5290 | PIK3CA, CLOVE, CWS5, MCAP, MCM, MCMTC, PI3K, p110-alpha | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (EC:2.7.11.1 2.7.1.153) |
C00044491
|
| 29990 | PILRB, FDFACT1, FDFACT2 | paired immunoglobin-like type 2 receptor beta |
C00044491
|
| 5292 | PIM1, PIM | pim-1 oncogene (EC:2.7.11.1) |
C00044491
|
| 5300 | PIN1, DOD, UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 (EC:5.2.1.8) |
C00044491
|
| 65018 | PINK1, BRPK, PARK6 | PTEN induced putative kinase 1 (EC:2.7.11.1) |
C00044491
|
| 5305 | PIP4K2A, PI5P4KA, PIP5K2A, PIP5KII-alpha, PIP5KIIA, PIPK | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha (EC:2.7.1.149) |
C00044491
|
| 79837 | PIP4K2C, PIP5K2C | phosphatidylinositol-5-phosphate 4-kinase, type II, gamma (EC:2.7.1.149) |
C00044491
|
| 8395 | PIP5K1B, MSS4, STM7 | phosphatidylinositol-4-phosphate 5-kinase, type I, beta (EC:2.7.1.68) |
C00044491
|
| 57095 | PITHD1, C1orf128, HT014, RP5-886K2.4, TXNL1CL | PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
C00044491
|
| 5306 | PITPNA, PI-TPalpha, PITPN, VIB1A | phosphatidylinositol transfer protein, alpha |
C00044491
|
| 26207 | PITPNC1, M-RDGB-beta, MRDGBbeta, RDGB-BETA, RDGBB, RDGBB1 | phosphatidylinositol transfer protein, cytoplasmic 1 |
C00044491
|
| 9600 | PITPNM1, DRES9, NIR2, PITPNM, RDGB, RDGB1, RDGBA, RDGBA1, Rd9 | phosphatidylinositol transfer protein, membrane-associated 1 |
C00044491
|
| 10531 | PITRM1, MP1, PreP | pitrilysin metallopeptidase 1 |
C00044491
|
| 168507 | PKD1L1, PRO19563 | polycystic kidney disease 1 like 1 |
C00044491
|
| 5570 | PKIB, PRKACN2 | protein kinase (cAMP-dependent, catalytic) inhibitor beta |
C00044491
|
| 5313 | PKLR, PK1, PKL, PKR, PKRL, RPK | pyruvate kinase, liver and RBC (EC:2.7.1.40) |
C00044491
|
| 9088 | PKMYT1, MYT1 | protein kinase, membrane associated tyrosine/threonine 1 (EC:2.7.11.1) |
C00044491
|
| 5316 | PKNOX1, PREP1, pkonx1c | PBX/knotted 1 homeobox 1 |
C00044491
|
| 5318 | PKP2, ARVD9 | plakophilin 2 |
C00044491
|
| 8502 | PKP4, p0071 | plakophilin 4 |
C00044491
|
| 81579 | PLA2G12A, GXII, PLA2G12, ROSSY | phospholipase A2, group XIIA (EC:3.1.1.4) |
C00044491
|
| 23659 | PLA2G15, ACS, GXVPLA2, LLPL, LPLA2, LYPLA3 | phospholipase A2, group XV (EC:2.3.1.-) |
C00044491
|
| 26279 | PLA2G2D, PLA2IID, SPLASH, sPLA2-IID, sPLA2S | phospholipase A2, group IID (EC:3.1.1.4) |
C00044491
|
| 8605 | PLA2G4C, CPLA2-gamma | phospholipase A2, group IVC (cytosolic, calcium-independent) (EC:3.1.1.4) |
C00044491
|
| 5335 | PLCG1, NCKAP3, PLC-II, PLC1, PLC148, PLCgamma1 | phospholipase C, gamma 1 (EC:3.1.4.11) |
C00044491
|
| 23228 | PLCL2, PLCE2 | phospholipase C-like 2 |
C00044491
|
| 5337 | PLD1 | phospholipase D1, phosphatidylcholine-specific (EC:3.1.4.4) |
C00044491
|
| 23646 | PLD3, HU-K4, HUK4 | phospholipase D family, member 3 (EC:3.1.4.4) |
C00044491
|
| 201164 | PLD6, ZUC | phospholipase D family, member 6 (EC:3.1.4.4) |
C00044491
|
| 5339 | PLEC, EBS1, EBSO, HD1, LGMD2Q, PCN, PLEC1, PLEC1b, PLTN | plectin |
C00044491
|
| 26499 | PLEK2 | pleckstrin 2 |
C00044491
|
| 59338 | PLEKHA1, TAPP1 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
C00044491
|
| 54477 | PLEKHA5, PEPP-2, PEPP2 | pleckstrin homology domain containing, family A member 5 |
C00044491
|
| 22874 | PLEKHA6, PEPP-3, PEPP3 | pleckstrin homology domain containing, family A member 6 |
C00044491
|
| 79666 | PLEKHF2, EAPF, PHAFIN2, ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
C00044491
|
| 26030 | PLEKHG3, ARHGEF43, KIAA0599 | pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
C00044491
|
| 57475 | PLEKHH1 | pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
C00044491
|
| 130271 | PLEKHH2, PLEKHH1L | pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
C00044491
|
| 9842 | PLEKHM1, AP162, B2, OPTB6 | pleckstrin homology domain containing, family M (with RUN domain) member 1 |
C00044491
|
| 23207 | PLEKHM2, RP11-169K16.1, SKIP | pleckstrin homology domain containing, family M (with RUN domain) member 2 |
C00044491
|
| 5340 | PLG | plasminogen (EC:3.4.21.7) |
C00044491
|
| 55848 | PLGRKT, C9orf46, MDS030, PLG-RKT, Plg-R(KT) | plasminogen receptor, C-terminal lysine transmembrane protein |
C00044491
|
| 123 | PLIN2, ADFP, ADRP | perilipin 2 |
C00044491
|
| 10226 | PLIN3, M6PRBP1, PP17, TIP47 | perilipin 3 |
C00044491
|
| 440503 | PLIN5, LSDA5, LSDP5, MLDP, OXPAT | perilipin 5 |
C00044491
|
| 1263 | PLK3, CNK, FNK, PRK | polo-like kinase 3 (EC:2.7.11.21) |
C00044491
|
| 10733 | PLK4, SAK, STK18 | polo-like kinase 4 (EC:2.7.11.21) |
C00044491
|
| 5351 | PLOD1, EDS6, LH, LH1, LLH, PLOD | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 (EC:1.14.11.4) |
C00044491
|
| 5352 | PLOD2, LH2, TLH | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (EC:1.14.11.4) |
C00044491
|
| 8985 | PLOD3, LH3 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (EC:1.14.11.4) |
C00044491
|
| 5355 | PLP2, A4, A4LSB | proteolipid protein 2 (colonic epithelium-enriched) |
C00044491
|
| 5357 | PLS1 | plastin 1 |
C00044491
|
| 5358 | PLS3, BMND18, T-plastin | plastin 3 |
C00044491
|
| 5359 | PLSCR1, MMTRA1B | phospholipid scramblase 1 |
C00044491
|
| 57088 | PLSCR4, TRA1 | phospholipid scramblase 4 |
C00044491
|
| 5360 | PLTP, BPIFE, HDLCQ9 | phospholipid transfer protein |
C00044491
|
| 83483 | PLVAP, FELS, PV-1, PV1, gp68 | plasmalemma vesicle associated protein |
C00044491
|
| 55558 | PLXNA3, 6.3, HSSEXGENE, PLXN3, PLXN4, XAP-6 | plexin A3 |
C00044491
|
| 5364 | PLXNB1, PLEXIN-B1, PLXN5, SEP | plexin B1 |
C00044491
|
| 23654 | PLXNB2, MM1, Nbla00445, PLEXB2, dJ402G11.3 | plexin B2 |
C00044491
|
| 135293 | PM20D2, ACY1L2, bA63L7.3 | peptidase M20 domain containing 2 |
C00044491
|
| 5366 | PMAIP1, APR, NOXA | phorbol-12-myristate-13-acetate-induced protein 1 |
C00044491
|
| 6490 | PMEL, D12S53E, ME20, ME20-M, ME20M, P1, P100, PMEL17, SI, SIL, SILV, gp100 | premelanosome protein |
C00044491
|
| 5378 | PMS1, HNPCC3, PMSL1, hPMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
C00044491
|
| 25957 | PNISR, C6orf111, HSPC306, RP11-98I9.2, SFRS18, SRrp130, bA98I9.2 | PNN-interacting serine/arginine-rich protein |
C00044491
|
| 25953 | PNKD, BRP17, DYT8, FPD1, KIPP1184, MR-1, MR1, PDC, PKND1, TAHCCP2 | paroxysmal nonkinesigenic dyskinesia (EC:3.1.2.6) |
C00044491
|
| 29944 | PNMA3, MA3, MA5 | paraneoplastic Ma antigen 3 |
C00044491
|
| 4860 | PNP, NP, PRO1837, PUNP | purine nucleoside phosphorylase (EC:2.4.2.1) |
C00044491
|
| 80339 | PNPLA3, ADPN, C22orf20, iPLA(2)epsilon | patatin-like phospholipase domain containing 3 (EC:3.1.1.3) |
C00044491
|
| 10908 | PNPLA6, NTE, NTEMND, SPG39, iPLA2delta, sws | patatin-like phospholipase domain containing 6 (EC:3.1.1.5) |
C00044491
|
| 375775 | PNPLA7, C9orf111, NTE-R1, NTEL1 | patatin-like phospholipase domain containing 7 |
C00044491
|
| 50640 | PNPLA8, IPLA2(GAMMA), IPLA2-2, IPLA2G, iPLA2gamma | patatin-like phospholipase domain containing 8 (EC:3.1.1.5) |
C00044491
|
| 87178 | PNPT1, COXPD13, DFNB70, OLD35, PNPASE, old-35 | polyribonucleotide nucleotidyltransferase 1 (EC:2.7.7.8) |
C00044491
|
| 25886 | POC1A, PIX2, SOFT, WDR51A | POC1 centriolar protein A |
C00044491
|
| 134359 | POC5, C5orf37 | POC5 centriolar protein |
C00044491
|
| 23126 | POGZ, ZNF280E, ZNF635, ZNF635m | pogo transposable element with ZNF domain |
C00044491
|
| 23649 | POLA2 | polymerase (DNA directed), alpha 2, accessory subunit |
C00044491
|
| 10714 | POLD3, P66, P68 | polymerase (DNA-directed), delta 3, accessory subunit |
C00044491
|
| 5426 | POLE, CRCS12, FILS, POLE1 | polymerase (DNA directed), epsilon, catalytic subunit (EC:2.7.7.7) |
C00044491
|
| 5427 | POLE2, DPE2 | polymerase (DNA directed), epsilon 2, accessory subunit (EC:2.7.7.7) |
C00044491
|
| 5428 | POLG, MDP1, MIRAS, MTDPS4A, MTDPS4B, PEO, POLG1, POLGA, SANDO, SCAE | polymerase (DNA directed), gamma (EC:2.7.7.7) |
C00044491
|
| 11232 | POLG2, HP55, MTPOLB, PEOA4, POLB, POLG-BETA, POLGB | polymerase (DNA directed), gamma 2, accessory subunit (EC:2.7.7.7) |
C00044491
|
| 5429 | POLH, RAD30, RAD30A, XP-V, XPV | polymerase (DNA directed), eta (EC:2.7.7.7) |
C00044491
|
| 10721 | POLQ, POLH, PRO0327 | polymerase (DNA directed), theta (EC:2.7.7.7) |
C00044491
|
| 84172 | POLR1B, RPA135, RPA2, Rpo1-2 | polymerase (RNA) I polypeptide B, 128kDa (EC:2.7.7.6) |
C00044491
|
| 9533 | POLR1C, RPA39, RPA40, RPA5, RPAC1, TCS3 | polymerase (RNA) I polypeptide C, 30kDa |
C00044491
|
| 51082 | POLR1D, AC19, POLR1C, RPA16, RPA9, RPAC2, RPC16, RPO1-3, TCS2 | polymerase (RNA) I polypeptide D, 16kDa |
C00044491
|
| 5430 | POLR2A, POLR2, POLRA, RPB1, RPBh1, RPO2, RPOL2, RpIILS, hRPB220, hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa (EC:2.7.7.6 2.7.7.48) |
C00044491
|
| 5433 | POLR2D, HSRBP4, HSRPB4, RBP4, RPB16 | polymerase (RNA) II (DNA directed) polypeptide D (EC:2.7.7.6) |
C00044491
|
| 5434 | POLR2E, RPABC1, RPB5, XAP4, hRPB25, hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa (EC:2.7.7.6) |
C00044491
|
| 55703 | POLR3B, C128, HLD8, RPC2 | polymerase (RNA) III (DNA directed) polypeptide B (EC:2.7.7.6) |
C00044491
|
| 5442 | POLRMT, APOLMT, MTRNAP, MTRPOL, h-mtRPOL | polymerase (RNA) mitochondrial (DNA directed) (EC:2.7.7.6) |
C00044491
|
| 5443 | POMC, ACTH, CLIP, LPH, MSH, NPP, POC | proopiomelanocortin |
C00044491
|
| 55624 | POMGNT1, GNTI.2, GnT_I.2, LGMD2O, MDDGA3, MDDGB3, MDDGC3, MEB, MGAT1.2, gnT-I.2 | protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) (EC:2.4.1.-) |
C00044491
|
| 5446 | PON3 | paraoxonase 3 (EC:3.1.1.2 3.1.8.1 3.1.1.81) |
C00044491
|
| 5447 | POR, CPR, CYPOR, P450R | P450 (cytochrome) oxidoreductase (EC:1.6.2.4) |
C00044491
|
| 64840 | PORCN, DHOF, FODH, MG61, PORC, PPN | porcupine homolog (Drosophila) |
C00044491
|
| 10631 | POSTN, OSF-2, OSF2, PDLPOSTN, PN, periostin | periostin, osteoblast specific factor |
C00044491
|
| 445582 | POTEE, A26C1, A26C1A, CT104.2, POTE-2, POTE2, POTE2gamma | POTE ankyrin domain family, member E |
C00044491
|
| 5464 | PPA1, IOPPP, PP, PP1, SID6-8061 | pyrophosphatase (inorganic) 1 (EC:3.6.1.1) |
C00044491
|
| 8613 | PPAP2B, Dri42, LPP3, PAP2B, VCIP | phosphatidic acid phosphatase type 2B (EC:3.1.3.4) |
C00044491
|
| 403313 | PPAPDC2, PDP1, PSDP, bA6J24.6 | phosphatidic acid phosphatase type 2 domain containing 2 |
C00044491
|
| 5465 | PPARA, NR1C1, PPAR, PPARalpha, hPPAR | peroxisome proliferator-activated receptor alpha |
C00044491
|
| 5467 | PPARD, FAAR, NR1C2, NUC1, NUCI, NUCII, PPARB | peroxisome proliferator-activated receptor delta |
C00044491
|
| 8500 | PPFIA1, LIP.1, LIP1, LIPRIN | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
C00044491
|
| 5481 | PPID, CYP-40, CYPD | peptidylprolyl isomerase D (EC:5.2.1.8) |
C00044491
|
| 51645 | PPIL1, CYPL1, PPIase, hCyPX | peptidylprolyl isomerase (cyclophilin)-like 1 (EC:5.2.1.8) |
C00044491
|
| 53938 | PPIL3, CYPJ | peptidylprolyl isomerase (cyclophilin)-like 3 (EC:5.2.1.8) |
C00044491
|
| 85313 | PPIL4, HDCME13P | peptidylprolyl isomerase (cyclophilin)-like 4 (EC:5.2.1.8) |
C00044491
|
| 23262 | PPIP5K2, HISPPD1, IP7K2, VIP2 | diphosphoinositol pentakisphosphate kinase 2 (EC:2.7.4.21 2.7.4.24) |
C00044491
|
| 5493 | PPL | periplakin |
C00044491
|
| 5495 | PPM1B, PP2C-beta-X, PP2CB, PP2CBETA, PPC2BETAX | protein phosphatase, Mg2+/Mn2+ dependent, 1B (EC:3.1.3.16) |
C00044491
|
| 8493 | PPM1D, PP2C-DELTA, WIP1 | protein phosphatase, Mg2+/Mn2+ dependent, 1D (EC:3.1.3.16) |
C00044491
|
| 5514 | PPP1R10, CAT53, FB19, PNUTS, PP1R10, R111, p99 | protein phosphatase 1, regulatory subunit 10 |
C00044491
|
| 26472 | PPP1R14B, PHI-1, PLCB3N, PNG, SOM172 | protein phosphatase 1, regulatory (inhibitor) subunit 14B |
C00044491
|
| 23645 | PPP1R15A, GADD34 | protein phosphatase 1, regulatory subunit 15A |
C00044491
|
| 84988 | PPP1R16A, MYPT3 | protein phosphatase 1, regulatory subunit 16A |
C00044491
|
| 170954 | PPP1R18, HKMT1098, KIAA1949 | protein phosphatase 1, regulatory subunit 18 |
C00044491
|
| 5502 | PPP1R1A, I1, IPP1 | protein phosphatase 1, regulatory (inhibitor) subunit 1A (EC:3.1.3.16) |
C00044491
|
| 79660 | PPP1R3B, GL, PPP1R4, PTG | protein phosphatase 1, regulatory subunit 3B |
C00044491
|
| 5507 | PPP1R3C, PPP1R5 | protein phosphatase 1, regulatory subunit 3C |
C00044491
|
| 5510 | PPP1R7, SDS22 | protein phosphatase 1, regulatory subunit 7 |
C00044491
|
| 5511 | PPP1R8, ARD-1, ARD1, NIPP-1, NIPP1, PRO2047 | protein phosphatase 1, regulatory subunit 8 |
C00044491
|
| 5515 | PPP2CA, PP2Ac, PP2CA, PP2Calpha, RP-C | protein phosphatase 2, catalytic subunit, alpha isozyme (EC:3.1.3.16) |
C00044491
|
| 28227 | PPP2R3B, NYREN8, PPP2R3L, PPP2R3LY, PR48 | protein phosphatase 2, regulatory subunit B'', beta |
C00044491
|
| 5524 | PPP2R4, PP2A, PR53, PTPA | protein phosphatase 2A activator, regulatory subunit 4 (EC:5.2.1.8) |
C00044491
|
| 5528 | PPP2R5D, B56D | protein phosphatase 2, regulatory subunit B', delta |
C00044491
|
| 5529 | PPP2R5E | protein phosphatase 2, regulatory subunit B', epsilon isoform |
C00044491
|
| 5530 | PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B | protein phosphatase 3, catalytic subunit, alpha isozyme (EC:3.1.3.16) |
C00044491
|
| 9989 | PPP4R1, MEG1, PP4(Rmeg), PP4R1 | protein phosphatase 4, regulatory subunit 1 |
C00044491
|
| 151987 | PPP4R2, PP4R2 | protein phosphatase 4, regulatory subunit 2 |
C00044491
|
| 57718 | PPP4R4, KIAA1622, PP4R4 | protein phosphatase 4, regulatory subunit 4 |
C00044491
|
| 55291 | PPP6R3, C11orf23, PP6R3, SAP190, SAPL, SAPLa, SAPS3 | protein phosphatase 6, regulatory subunit 3 |
C00044491
|
| 23082 | PPRC1, PRC, RP11-302K17.6 | peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
C00044491
|
| 5538 | PPT1, CLN1, INCL, PPT | palmitoyl-protein thioesterase 1 (EC:3.1.2.22) |
C00044491
|
| 160760 | PPTC7, TA-PP2C, TAPP2C | PTC7 protein phosphatase homolog (S. cerevisiae) (EC:3.1.3.16) |
C00044491
|
| 54896 | PQLC2 | PQ loop repeat containing 2 |
C00044491
|
| 343071 | PRAMEF10, RP5-845O24.7 | PRAME family member 10 |
C00044491
|
| 9055 | PRC1, ASE1 | protein regulator of cytokinesis 1 |
C00044491
|
| 56980 | PRDM10, PFM7 | PR domain containing 10 |
C00044491
|
| 59336 | PRDM13, MU-MB-20.220, PFM10 | PR domain containing 13 |
C00044491
|
| 63976 | PRDM16, CMD1LL, LVNC8, MEL1, PFM13 | PR domain containing 16 |
C00044491
|
| 7799 | PRDM2, HUMHOXY1, KMT8, MTB-ZF, RIZ, RIZ1, RIZ2 | PR domain containing 2, with ZNF domain (EC:2.1.1.43) |
C00044491
|
| 10113 | PREB, SEC12 | prolactin regulatory element binding |
C00044491
|
| 27166 | PRELID1, PRELI, PX19, SBBI12 | PRELI domain containing 1 |
C00044491
|
| 153768 | PRELID2 | PRELI domain containing 2 |
C00044491
|
| 9581 | PREPL | prolyl endopeptidase-like (EC:3.4.21.-) |
C00044491
|
| 5557 | PRIM1, p49 | primase, DNA, polypeptide 1 (49kDa) (EC:2.7.7.7) |
C00044491
|
| 5562 | PRKAA1, AMPK, AMPKa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit (EC:2.7.11.1 2.7.11.26 2.7.11.27 2.7.11.31) |
C00044491
|
| 5565 | PRKAB2 | protein kinase, AMP-activated, beta 2 non-catalytic subunit |
C00044491
|
| 5576 | PRKAR2A, PKR2, PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha (EC:2.7.11.1) |
C00044491
|
| 5578 | PRKCA, AAG6, PKC-alpha, PKCA, PRKACA | protein kinase C, alpha (EC:2.7.11.13) |
C00044491
|
| 5579 | PRKCB, PKC-beta, PKCB, PRKCB1, PRKCB2 | protein kinase C, beta (EC:2.7.11.13) |
C00044491
|
| 5583 | PRKCH, PKC-L, PKCL, PRKCL, nPKC-eta | protein kinase C, eta (EC:2.7.11.13) |
C00044491
|
| 5588 | PRKCQ, PRKCT, nPKC-theta | protein kinase C, theta (EC:2.7.11.13) |
C00044491
|
| 5589 | PRKCSH, AGE-R2, G19P1, PCLD, PKCSH, PLD1 | protein kinase C substrate 80K-H (EC:2.7.11.1) |
C00044491
|
| 5590 | PRKCZ, PKC-ZETA, PKC2 | protein kinase C, zeta (EC:2.7.11.13) |
C00044491
|
| 23683 | PRKD3, EPK2, PKC-NU, PKD3, PRKCN, nPKC-NU | protein kinase D3 (EC:2.7.11.13) |
C00044491
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00044491
|
| 8575 | PRKRA, DYT16, PACT, RAX | protein kinase, interferon-inducible double stranded RNA dependent activator |
C00044491
|
| 58531 | PRM3 | protamine 3 |
C00044491
|
| 10419 | PRMT5, HRMT1L5, IBP72, JBP1, SKB1, SKB1Hs | protein arginine methyltransferase 5 (EC:2.1.1.125) |
C00044491
|
| 5621 | PRNP, ASCR, AltPrP, CD230, CJD, GSS, KURU, PRIP, PrP, PrP27-30, PrP33-35C, PrPc, p27-30 | prion protein |
C00044491
|
| 147011 | PROCA1 | protein interacting with cyclin A1 |
C00044491
|
| 58510 | PRODH2, HSPOX1 | proline dehydrogenase (oxidase) 2 (EC:1.5.99.8) |
C00044491
|
| 5627 | PROS1, PROS, PS21, PS22, PS23, PS24, PS25, PSA, THPH5, THPH6 | protein S (alpha) |
C00044491
|
| 5629 | PROX1 | prospero homeobox 1 |
C00044491
|
| 55015 | PRPF39 | pre-mRNA processing factor 39 |
C00044491
|
| 8899 | PRPF4B, PR4H, PRP4, PRP4H, PRP4K, dJ1013A10.1 | pre-mRNA processing factor 4B (EC:2.7.11.1) |
C00044491
|
| 5635 | PRPSAP1, PAP39 | phosphoribosyl pyrophosphate synthetase-associated protein 1 |
C00044491
|
| 5636 | PRPSAP2, PAP41 | phosphoribosyl pyrophosphate synthetase-associated protein 2 |
C00044491
|
| 55771 | PRR11 | proline rich 11 |
C00044491
|
| 7916 | PRRC2A, BAT2, D6S51, D6S51E, G2 | proline-rich coiled-coil 2A |
C00044491
|
| 23215 | PRRC2C, BAT2-iso, BAT2D1, BAT2L2, XTP2 | proline-rich coiled-coil 2C |
C00044491
|
| 83886 | PRSS27, CAPH2, MPN | protease, serine 27 |
C00044491
|
| 360226 | PRSS41, TESSP1 | protease, serine, 41 (EC:3.4.21.-) |
C00044491
|
| 5657 | PRTN3, ACPA, AGP7, C-ANCA, CANCA, MBN, MBT, NP-4, NP4, P29, PR-3, PR3 | proteinase 3 (EC:3.4.21.76) |
C00044491
|
| 414327 |
C00044491
|
||
| 5660 | PSAP, GLBA, SAP1 | prosaposin |
C00044491
|
| 29968 | PSAT1, EPIP, PSA, PSAT | phosphoserine aminotransferase 1 (EC:2.6.1.52) |
C00044491
|
| 23362 | PSD3, EFA6R, HCA67 | pleckstrin and Sec7 domain containing 3 |
C00044491
|
| 23550 | PSD4, EFA6B, TIC | pleckstrin and Sec7 domain containing 4 |
C00044491
|
| 11168 | PSIP1, DFS70, LEDGF, PAIP, PSIP2, p52, p75 | PC4 and SFRS1 interacting protein 1 |
C00044491
|
| 5684 | PSMA3, HC8, PSC3 | proteasome (prosome, macropain) subunit, alpha type, 3 (EC:3.4.25.1) |
C00044491
|
| 5699 | PSMB10, LMP10, MECL1, beta2i | proteasome (prosome, macropain) subunit, beta type, 10 (EC:3.4.25.1) |
C00044491
|
| 5690 | PSMB2, HC7-I | proteasome (prosome, macropain) subunit, beta type, 2 (EC:3.4.25.1) |
C00044491
|
| 5695 | PSMB7, Z | proteasome (prosome, macropain) subunit, beta type, 7 (EC:3.4.25.1) |
C00044491
|
| 5696 | PSMB8, ALDD, D6S216, D6S216E, JMP, LMP7, NKJO, PSMB5i, RING10 | proteasome (prosome, macropain) subunit, beta type, 8 (EC:3.4.25.1) |
C00044491
|
| 5698 | PSMB9, LMP2, PSMB6i, RING12, beta1i | proteasome (prosome, macropain) subunit, beta type, 9 (EC:3.4.25.1) |
C00044491
|
| 5701 | PSMC2, MSS1, Nbla10058, S7 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
C00044491
|
| 5704 | PSMC4, MIP224, RPT3, S6, TBP-7, TBP7 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
C00044491
|
| 5718 | PSMD12, Rpn5, p55 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
C00044491
|
| 5711 | PSMD5, S5B | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
C00044491
|
| 10197 | PSME3, Ki, PA28-gamma, PA28G, PA28gamma, REG-GAMMA | proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
C00044491
|
| 9491 | PSMF1, PI31 | proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
C00044491
|
| 8624 | PSMG1, C21LRP, DSCR2, LRPC21, PAC-1, PAC1 | proteasome (prosome, macropain) assembly chaperone 1 |
C00044491
|
| 56984 | PSMG2, CLAST3, HCCA3, HsT1707, MDS003, PAC2, TNFSF5IP1 | proteasome (prosome, macropain) assembly chaperone 2 |
C00044491
|
| 389362 | PSMG4, C6orf86, PAC4, bA506K6.2 | proteasome (prosome, macropain) assembly chaperone 4 |
C00044491
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00044491
|
| 9050 | PSTPIP2, MAYP | proline-serine-threonine phosphatase interacting protein 2 |
C00044491
|
| 5724 | PTAFR, PAFR | platelet-activating factor receptor |
C00044491
|
| 442213 | PTCHD4, C6orf138, dJ402H5.2 | patched domain containing 4 |
C00044491
|
| 5738 | PTGFRN, CD315, CD9P-1, EWI-F, FPRP, SMAP-6 | prostaglandin F2 receptor inhibitor |
C00044491
|
| 22949 | PTGR1, LTB4DH, PGR1, ZADH3 | prostaglandin reductase 1 (EC:1.3.1.48 1.3.1.74) |
C00044491
|
| 145482 | PTGR2, PGR2, ZADH1 | prostaglandin reductase 2 (EC:1.3.1.48) |
C00044491
|
| 5742 | PTGS1, COX1, COX3, PCOX1, PES-1, PGG/HS, PGHS-1, PGHS1, PHS1, PTGHS | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00044491
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00044491
|
| 113091 | PTH2, TIP39 | parathyroid hormone 2 |
C00044491
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00044491
|
| 2185 | PTK2B, CADTK, CAKB, FADK2, FAK2, PKB, PTK, PYK2, RAFTK | protein tyrosine kinase 2 beta (EC:2.7.10.2) |
C00044491
|
| 7803 | PTP4A1, HH72, PRL-1, PRL1, PTP(CAAX1), PTPCAAX1 | protein tyrosine phosphatase type IVA, member 1 (EC:3.1.3.48) |
C00044491
|
| 138639 | PTPDC1, PTP9Q22 | protein tyrosine phosphatase domain containing 1 (EC:3.1.3.48) |
C00044491
|
| 51495 | PTPLAD1, B-IND1, HACD3, HSPC121 | protein tyrosine phosphatase-like A domain containing 1 (EC:4.2.1.134) |
C00044491
|
| 5781 | PTPN11, BPTP3, CFC, NS1, PTP-1D, PTP2C, SH-PTP2, SH-PTP3, SHP2 | protein tyrosine phosphatase, non-receptor type 11 (EC:3.1.3.48) |
C00044491
|
| 5774 | PTPN3, PTP-H1, PTPH1 | protein tyrosine phosphatase, non-receptor type 3 (EC:3.1.3.48) |
C00044491
|
| 5792 | PTPRF, LAR | protein tyrosine phosphatase, receptor type, F (EC:3.1.3.48) |
C00044491
|
| 5795 | PTPRJ, CD148, DEP1, HPTPeta, R-PTP-ETA, SCC1 | protein tyrosine phosphatase, receptor type, J (EC:3.1.3.48) |
C00044491
|
| 5796 | PTPRK, R-PTP-kappa | protein tyrosine phosphatase, receptor type, K (EC:3.1.3.48) |
C00044491
|
| 10076 | PTPRU, FMI, PCP-2, PTP, PTP-J, PTP-PI, PTP-RO, PTPPSI, PTPRO, PTPU2, R-PTP-PSI, R-PTP-U, hPTP-J | protein tyrosine phosphatase, receptor type, U (EC:3.1.3.48) |
C00044491
|
| 9232 | PTTG1, EAP1, HPTTG, PTTG, TUTR1 | pituitary tumor-transforming 1 |
C00044491
|
| 23369 | PUM2, PUMH2, PUML2 | pumilio RNA-binding family member 2 |
C00044491
|
| 150962 | PUS10, CCDC139, DOBI | pseudouridylate synthase 10 |
C00044491
|
| 5819 | PVRL2, CD112, HVEB, PRR2, PVRR2 | poliovirus receptor-related 2 (herpesvirus entry mediator B) |
C00044491
|
| 81607 | PVRL4, EDSS1, LNIR, PRR4, nectin-4 | poliovirus receptor-related 4 |
C00044491
|
| 11137 | PWP1, IEF-SSP-9502 | PWP1 homolog (S. cerevisiae) |
C00044491
|
| 54899 | PXK, MONaKA | PX domain containing serine/threonine kinase |
C00044491
|
| 5829 | PXN | paxillin |
C00044491
|
| 5831 | PYCR1, ARCL2B, ARCL3B, P5C, P5CR, PIG45, PP222, PRO3, PYCR | pyrroline-5-carboxylate reductase 1 (EC:1.5.1.2) |
C00044491
|
| 5836 | PYGL, GSD6 | phosphorylase, glycogen, liver (EC:2.4.1.1) |
C00044491
|
| 347148 | QRFP, 26RFa, P518 | pyroglutamylated RFamide peptide |
C00044491
|
| 79832 | QSER1 | glutamine and serine rich 1 |
C00044491
|
| 169714 | QSOX2, QSCN6L1, SOXN | quiescin Q6 sulfhydryl oxidase 2 (EC:1.8.3.2) |
C00044491
|
| 79691 | QTRTD1 | queuine tRNA-ribosyltransferase domain containing 1 (EC:2.4.2.29) |
C00044491
|
| 91300 | R3HDM4, C19orf22 | R3H domain containing 4 |
C00044491
|
| 10890 | RAB10 | RAB10, member RAS oncogene family |
C00044491
|
| 9727 | RAB11FIP3, CART1, Rab11-FIP3 | RAB11 family interacting protein 3 (class II) (EC:2.7.4.6) |
C00044491
|
| 22931 | RAB18, RAB18LI1, WARBM3 | RAB18, member RAS oncogene family |
C00044491
|
| 5861 | RAB1A, RAB1, YPT1 | RAB1A, member RAS oncogene family |
C00044491
|
| 55647 | RAB20 | RAB20, member RAS oncogene family |
C00044491
|
| 5873 | RAB27A, GS2, HsT18676, RAB27, RAM | RAB27A, member RAS oncogene family |
C00044491
|
| 84932 | RAB2B | RAB2B, member RAS oncogene family |
C00044491
|
| 83452 | RAB33B, SMC2 | RAB33B, member RAS oncogene family |
C00044491
|
| 11021 | RAB35, H-ray, RAB1C, RAY | RAB35, member RAS oncogene family |
C00044491
|
| 23682 | RAB38, NY-MEL-1, rrGTPbp | RAB38, member RAS oncogene family |
C00044491
|
| 5864 | RAB3A | RAB3A, member RAS oncogene family |
C00044491
|
| 25782 | RAB3GAP2, RAB3-GAP150, RAB3GAP150, WARBM2, p150 | RAB3 GTPase activating protein subunit 2 (non-catalytic) |
C00044491
|
| 5870 | RAB6A, RAB6 | RAB6A, member RAS oncogene family |
C00044491
|
| 8934 | RAB7L1, RAB7L | RAB7, member RAS oncogene family-like 1 |
C00044491
|
| 23637 | RABGAP1, GAPCENA, RP11-123N4.2, TBC1D11 | RAB GTPase activating protein 1 |
C00044491
|
| 9910 | RABGAP1L, HHL, TBC1D18 | RAB GTPase activating protein 1-like |
C00044491
|
| 27342 | RABGEF1, RABEX5, rabex-5 | RAB guanine nucleotide exchange factor (GEF) 1 |
C00044491
|
| 5875 | RABGGTA, PTAR3 | Rab geranylgeranyltransferase, alpha subunit (EC:2.5.1.60) |
C00044491
|
| 5877 | RABIF, MSS4, RASGFR3, RASGRF3 | RAB interacting factor |
C00044491
|
| 29127 | RACGAP1, CYK4, HsCYK-4, ID-GAP, MgcRacGAP | Rac GTPase activating protein 1 |
C00044491
|
| 83956 | RACGAP1P, FKSG42 | Rac GTPase activating protein 1 pseudogene |
C00044491
|
| 56852 | RAD18, RNF73 | RAD18 homolog (S. cerevisiae) |
C00044491
|
| 10111 | RAD50, NBSLD, RAD502, hRad50 | RAD50 homolog (S. cerevisiae) |
C00044491
|
| 5888 | RAD51, BRCC5, HRAD51, HsRad51, HsT16930, MRMV2, RAD51A, RECA | RAD51 recombinase |
C00044491
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00044491
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00044491
|
| 25788 | RAD54B, RDH54 | RAD54 homolog B (S. cerevisiae) |
C00044491
|
| 8438 | RAD54L, HR54, RAD54A, hHR54, hRAD54 | RAD54-like (S. cerevisiae) |
C00044491
|
| 135250 | RAET1E, LETAL, N2DL-4, NKG2DL4, RAET1E2, RL-4, ULBP4, bA350J20.7 | retinoic acid early transcript 1E |
C00044491
|
| 26064 | RAI14, NORPEG, RAI13 | retinoic acid induced 14 |
C00044491
|
| 57186 | RALGAPA2, AS250, C20orf74, bA287B20.1, dJ1049G11, dJ1049G11.4, p220 | Ral GTPase activating protein, alpha subunit 2 (catalytic) |
C00044491
|
| 57148 | RALGAPB, KIAA1219, RalGAPbeta | Ral GTPase activating protein, beta subunit (non-catalytic) |
C00044491
|
| 5900 | RALGDS, RGDS, RGF, RalGEF | ral guanine nucleotide dissociation stimulator |
C00044491
|
| 5901 | RAN, ARA24, Gsp1, TC4 | RAN, member RAS oncogene family |
C00044491
|
| 57610 | RANBP10 | RAN binding protein 10 |
C00044491
|
| 5903 | RANBP2, ADANE, ANE1, IIAE3, NUP358, TRP1, TRP2 | RAN binding protein 2 (EC:5.2.1.8) |
C00044491
|
| 26953 | RANBP6 | RAN binding protein 6 |
C00044491
|
| 5905 | RANGAP1, Fug1, RANGAP, SD | Ran GTPase activating protein 1 |
C00044491
|
| 5909 | RAP1GAP, RAP1GA1, RAP1GAP1, RAP1GAPII, RAPGAP | RAP1 GTPase activating protein |
C00044491
|
| 5912 | RAP2B | RAP2B, member of RAS oncogene family |
C00044491
|
| 57826 | RAP2C | RAP2C, member of RAS oncogene family |
C00044491
|
| 65059 | RAPH1, ALS2CR18, ALS2CR9, LPD, PREL-2, PREL2, RMO1, RalGDS/AF-6 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
C00044491
|
| 5918 | RARRES1, LXNL, TIG1 | retinoic acid receptor responder (tazarotene induced) 1 |
C00044491
|
| 5919 | RARRES2, HP10433, TIG2 | retinoic acid receptor responder (tazarotene induced) 2 |
C00044491
|
| 5921 | RASA1, CM-AVM, CMAVM, GAP, PKWS, RASA, RASGAP, p120GAP, p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 |
C00044491
|
| 10125 | RASGRP1, CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
C00044491
|
| 11186 | RASSF1, 123F2, NORE2A, RASSF1A, RDA32, REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 |
C00044491
|
| 11228 | RASSF8, C12orf2, HOJ1 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
C00044491
|
| 30062 | RAX, MCOP3, RX | retina and anterior neural fold homeobox |
C00044491
|
| 84839 | RAX2, ARMD6, CORD11, QRX, RAXL1 | retina and anterior neural fold homeobox 2 |
C00044491
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00044491
|
| 5928 | RBBP4, NURF55, RBAP48 | retinoblastoma binding protein 4 |
C00044491
|
| 5929 | RBBP5, RBQ3, SWD1 | retinoblastoma binding protein 5 |
C00044491
|
| 5931 | RBBP7, RbAp46 | retinoblastoma binding protein 7 |
C00044491
|
| 10741 | RBBP9, BOG, RBBP10 | retinoblastoma binding protein 9 |
C00044491
|
| 64080 | RBKS, RBSK | ribokinase (EC:2.7.1.15) |
C00044491
|
| 5933 | RBL1, CP107, PRB1, p107 | retinoblastoma-like 1 (p107) |
C00044491
|
| 10432 | RBM14, COAA, PSP2, SIP, SYTIP1, TMEM137 | RNA binding motif protein 14 |
C00044491
|
| 64783 | RBM15, OTT, OTT1, SPEN | RNA binding motif protein 15 |
C00044491
|
| 64062 | RBM26, ARRS2, C13orf10, SE70-2, ZC3H17 | RNA binding motif protein 26 |
C00044491
|
| 55544 | RBM38, HSRNASEB, RNPC1, SEB4B, SEB4D, dJ800J21.2 | RNA binding motif protein 38 |
C00044491
|
| 9584 | RBM39, CAPER, CAPERalpha, FSAP59, HCC1, RNPC2 | RNA binding motif protein 39 |
C00044491
|
| 54502 | RBM47, NET18 | RNA binding motif protein 47 |
C00044491
|
| 10179 | RBM7 | RNA binding motif protein 7 |
C00044491
|
| 27316 | RBMX, HNRNPG, HNRPG, RBMXP1, RBMXRT, RNMX, hnRNP-G | RNA binding motif protein, X-linked |
C00044491
|
| 139804 | RBMXL3, CXorf55 | RNA binding motif protein, X-linked-like 3 |
C00044491
|
| 348093 | RBPMS2 | RNA binding protein with multiple splicing 2 |
C00044491
|
| 149041 | RC3H1, RNF198, ROQUIN | ring finger and CCCH-type domains 1 |
C00044491
|
| 1827 | RCAN1, ADAPT78, CSP1, DSC1, DSCR1, MCIP1, RCN1 | regulator of calcineurin 1 |
C00044491
|
| 55213 | RCBTB1, CLLD7, CLLL7, GLP, RP11-185C18.1 | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
C00044491
|
| 91433 | RCCD1 | RCC1 domain containing 1 |
C00044491
|
| 23186 | RCOR1, COREST, RCOR | REST corepressor 1 |
C00044491
|
| 283248 | RCOR2 | REST corepressor 2 |
C00044491
|
| 157506 | RDH10, SDR16C4 | retinol dehydrogenase 10 (all-trans) (EC:1.1.1.300) |
C00044491
|
| 8608 | RDH16, RODH-4, SDR9C8 | retinol dehydrogenase 16 (all-trans) |
C00044491
|
| 51308 | REEP2, C5orf19, SGC32445 | receptor accessory protein 2 |
C00044491
|
| 80346 | REEP4, C8orf20 | receptor accessory protein 4 |
C00044491
|
| 768211 | RELL1 | RELT-like 1 |
C00044491
|
| 5972 | REN, HNFJ2 | renin (EC:3.4.23.15) |
C00044491
|
| 11079 | RER1 | retention in endoplasmic reticulum sorting receptor 1 |
C00044491
|
| 473 | RERE, ARG, ARP, ATN1L, DNB1 | arginine-glutamic acid dipeptide (RE) repeats |
C00044491
|
| 54884 | RETSAT | retinol saturase (all-trans-retinol 13,14-reductase) (EC:1.3.99.23) |
C00044491
|
| 5980 | REV3L, POLZ, REV3 | REV3-like, polymerase (DNA directed), zeta, catalytic subunit (EC:2.7.7.7) |
C00044491
|
| 25996 | REXO2, REX2, RFN, SFN | RNA exonuclease 2 |
C00044491
|
| 5981 | RFC1, A1, MHCBFB, PO-GA, RECC1, RFC, RFC140 | replication factor C (activator 1) 1, 145kDa |
C00044491
|
| 5982 | RFC2, RFC40 | replication factor C (activator 1) 2, 40kDa |
C00044491
|
| 5983 | RFC3, RFC38 | replication factor C (activator 1) 3, 38kDa |
C00044491
|
| 5984 | RFC4, A1, RFC37 | replication factor C (activator 1) 4, 37kDa |
C00044491
|
| 5985 | RFC5, RFC36 | replication factor C (activator 1) 5, 36.5kDa |
C00044491
|
| 55312 | RFK, RIFK | riboflavin kinase (EC:2.7.1.26) |
C00044491
|
| 23180 | RFTN1, PIB10, PIG9, RAFTLIN | raftlin, lipid raft linker 1 |
C00044491
|
| 64326 | RFWD2, COP1, RNF200 | ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
C00044491
|
| 55159 | RFWD3, RNF201 | ring finger and WD repeat domain 3 |
C00044491
|
| 5993 | RFX5 | regulatory factor X, 5 (influences HLA class II expression) |
C00044491
|
| 28984 | RGCC, C13orf15, RGC-32, RGC32, bA157L14.2 | regulator of cell cycle |
C00044491
|
| 23179 | RGL1, RGL | ral guanine nucleotide dissociation stimulator-like 1 |
C00044491
|
| 9104 | RGN, GNL, RC, SMP30 | regucalcin (EC:3.1.1.17) |
C00044491
|
| 6002 | RGS12 | regulator of G-protein signaling 12 |
C00044491
|
| 10636 | RGS14 | regulator of G-protein signaling 14 |
C00044491
|
| 6004 | RGS16, A28-RGS14, A28-RGS14P, RGS-R | regulator of G-protein signaling 16 |
C00044491
|
| 5997 | RGS2, G0S8 | regulator of G-protein signaling 2, 24kDa |
C00044491
|
| 5998 | RGS3, C2PA, RGP3 | regulator of G-protein signaling 3 |
C00044491
|
| 57414 | RHBDD2, NPD007, RHBDL7 | rhomboid domain containing 2 |
C00044491
|
| 64285 | RHBDF1, C16orf8, Dist1, EGFR-RS, gene-89, gene-90, hDist1 | rhomboid 5 homolog 1 (Drosophila) |
C00044491
|
| 23221 | RHOBTB2, DBC2 | Rho-related BTB domain containing 2 |
C00044491
|
| 22836 | RHOBTB3 | Rho-related BTB domain containing 3 |
C00044491
|
| 23433 | RHOQ, ARHQ, RASL7A, TC10, TC10A | ras homolog family member Q |
C00044491
|
| 55288 | RHOT1, ARHT1, MIRO-1, MIRO1 | ras homolog family member T1 |
C00044491
|
| 58480 | RHOU, ARHU, CDC42L1, DJ646B12.2, WRCH1, fJ646B12.2, hG28K | ras homolog family member U |
C00044491
|
| 26150 | RIBC2, C22orf11 | RIB43A domain with coiled-coils 2 |
C00044491
|
| 60626 | RIC8A, RIC8 | RIC8 guanine nucleotide exchange factor A |
C00044491
|
| 253260 | RICTOR, AVO3, PIA, hAVO3 | RPTOR independent companion of MTOR, complex 2 |
C00044491
|
| 55183 | RIF1 | RAP1 interacting factor homolog (yeast) |
C00044491
|
| 54453 | RIN2, MACS, RASSF4 | Ras and Rab interactor 2 |
C00044491
|
| 126432 | RINL | Ras and Rab interactor-like |
C00044491
|
| 55781 | RIOK2, RIO2 | RIO kinase 2 (EC:2.7.11.1) |
C00044491
|
| 8780 | RIOK3, SUDD | RIO kinase 3 (EC:2.7.11.1) |
C00044491
|
| 54101 | RIPK4, ANKK2, ANKRD3, DIK, NKRD3, PKK, PPS2, RIP4 | receptor-interacting serine-threonine kinase 4 (EC:2.7.11.1) |
C00044491
|
| 134701 | RIPPLY2, C6orf159, dJ237I15.1 | ripply transcriptional repressor 2 |
C00044491
|
| 6018 | RLF, ZN-15L, ZNF292L | rearranged L-myc fusion |
C00044491
|
| 51115 | RMDN1, FAM82B, RMD-1, RMD1 | regulator of microtubule dynamics 1 |
C00044491
|
| 80010 | RMI1, BLAP75, C9orf76, FAAP75, RP11-346I8.1 | RecQ mediated genome instability 1 |
C00044491
|
| 64795 | RMND5A, CTLH, GID2, GID2A, RMD5, p44CTLH | required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
C00044491
|
| 6036 | RNASE2, EDN, RNS2 | ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (EC:3.1.27.5) |
C00044491
|
| 6038 | RNASE4, RNS4 | ribonuclease, RNase A family, 4 (EC:3.1.27.-) |
C00044491
|
| 567505 |
C00044491
|
||
| 27289 | RND1, ARHS, RHO6, RHOS | Rho family GTPase 1 |
C00044491
|
| 390 | RND3, ARHE, Rho8, RhoE, memB | Rho family GTPase 3 |
C00044491
|
| 55905 | RNF114, PSORS12, ZNF313 | ring finger protein 114 |
C00044491
|
| 63891 | RNF123, KPC1 | ring finger protein 123 |
C00044491
|
| 9604 | RNF14, ARA54, HFB30, TRIAD2 | ring finger protein 14 |
C00044491
|
| 50862 | RNF141, ZFP26, ZNF230 | ring finger protein 141 |
C00044491
|
| 255488 | RNF144B, IBRDC2, PIR2, bA528A10.3, p53RFP | ring finger protein 144B |
C00044491
|
| 165918 | RNF168, hRNF168 | ring finger protein 168, E3 ubiquitin protein ligase |
C00044491
|
| 56163 | RNF17, Mmip-2, SPATA23, TDRD4 | ring finger protein 17 |
C00044491
|
| 81790 | RNF170, SNAX1 | ring finger protein 170 |
C00044491
|
| 127544 | RNF19B, IBRDC3, NKLAM | ring finger protein 19B |
C00044491
|
| 6045 | RNF2, BAP-1, BAP1, DING, HIPI3, RING1B, RING2 | ring finger protein 2 (EC:6.3.2.-) |
C00044491
|
| 56254 | RNF20, BRE1, BRE1A, hBRE1 | ring finger protein 20, E3 ubiquitin protein ligase |
C00044491
|
| 57674 | RNF213, ALO17, C17orf27, KIAA1618, MYMY2, MYSTR, NET57 | ring finger protein 213 |
C00044491
|
| 79102 | RNF26 | ring finger protein 26 |
C00044491
|
| 80196 | RNF34, CARP-1, CARP1, RFI, RIF, RIFF, hRFI | ring finger protein 34, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00044491
|
| 152006 | RNF38 | ring finger protein 38 |
C00044491
|
| 9616 | RNF7, CKBBP1, ROC2, SAG | ring finger protein 7 |
C00044491
|
| 84900 | RNFT2, TMEM118 | ring finger protein, transmembrane 2 |
C00044491
|
| 6091 | ROBO1, DUTT1, SAX3 | roundabout, axon guidance receptor, homolog 1 (Drosophila) |
C00044491
|
| 54538 | ROBO4, ECSM4, MRB | roundabout, axon guidance receptor, homolog 4 (Drosophila) |
C00044491
|
| 6095 | RORA, NR1F1, ROR1, ROR2, ROR3, RZR-ALPHA, RZRA | RAR-related orphan receptor A |
C00044491
|
| 6097 | RORC, NR1F3, RORG, RZR-GAMMA, RZRG, TOR | RAR-related orphan receptor C |
C00044491
|
| 6117 | RPA1, HSSB, MST075, REPA1, RF-A, RP-A, RPA70 | replication protein A1, 70kDa |
C00044491
|
| 6118 | RPA2, REPA2, RPA32 | replication protein A2, 32kDa |
C00044491
|
| 6120 | RPE, RPE2-1 | ribulose-5-phosphate-3-epimerase (EC:5.1.3.1) |
C00044491
|
| 6136 | RPL12, L12 | ribosomal protein L12 |
C00044491
|
| 6137 | RPL13, BBC1, D16S444E, D16S44E, L13 | ribosomal protein L13 (EC:3.6.5.3) |
C00044491
|
| 9045 | RPL14, CAG-ISL-7, CTG-B33, L14, RL14, hRL14 | ribosomal protein L14 |
C00044491
|
| 6146 | RPL22, EAP, HBP15, HBP15/L22, L22 | ribosomal protein L22 |
C00044491
|
| 200916 | RPL22L1 | ribosomal protein L22-like 1 |
C00044491
|
| 644338 |
C00044491
|
||
| 6128 | RPL6, L6, SHUJUN-2, TAXREB107, TXREB1 | ribosomal protein L6 |
C00044491
|
| 6129 | RPL7, L7, humL7-1 | ribosomal protein L7 |
C00044491
|
| 6133 | RPL9, L9, NPC-A-16 | ribosomal protein L9 |
C00044491
|
| 10556 | RPP30, TSG15 | ribonuclease P/MRP 30kDa subunit (EC:3.1.26.5) |
C00044491
|
| 58490 | RPRD1B, C20orf77, CREPT, NET60, dJ1057B20.2 | regulation of nuclear pre-mRNA domain containing 1B |
C00044491
|
| 23248 | RPRD2, KIAA0460 | regulation of nuclear pre-mRNA domain containing 2 |
C00044491
|
| 56475 | RPRM, REPRIMO | reprimo, TP53 dependent G2 arrest mediator candidate |
C00044491
|
| 388394 | RPRML | reprimo-like |
C00044491
|
| 647190 | RPS16P5, RPS16_3_702 | ribosomal protein S16 pseudogene 5 |
C00044491
|
| 6228 | RPS23, S23 | ribosomal protein S23 |
C00044491
|
| 6229 | RPS24, DBA3, S24 | ribosomal protein S24 |
C00044491
|
| 6230 | RPS25, S25 | ribosomal protein S25 |
C00044491
|
| 51065 | RPS27L | ribosomal protein S27-like |
C00044491
|
| 6189 | RPS3A, FTE1, MFTL, S3A | ribosomal protein S3A |
C00044491
|
| 140032 | RPS4Y2, RPS4Y2P | ribosomal protein S4, Y-linked 2 |
C00044491
|
| 6195 | RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1) |
C00044491
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00044491
|
| 26750 | RPS6KC1, RPK118, humS6PKh1 | ribosomal protein S6 kinase, 52kDa, polypeptide 1 (EC:2.7.11.1) |
C00044491
|
| 6236 | RRAD, RAD, RAD1, REM3 | Ras-related associated with diabetes |
C00044491
|
| 64121 | RRAGC, GTR2, RAGC, TIB929 | Ras-related GTP binding C |
C00044491
|
| 6237 | RRAS | related RAS viral (r-ras) oncogene homolog |
C00044491
|
| 6239 | RREB1, FINB, HNT, LZ321, RREB-1, Zep-1 | ras responsive element binding protein 1 |
C00044491
|
| 6240 | RRM1, R1, RIR1, RR1 | ribonucleotide reductase M1 (EC:1.17.4.1) |
C00044491
|
| 50484 | RRM2B, MTDPS8A, MTDPS8B, P53R2 | ribonucleotide reductase M2 B (TP53 inducible) (EC:1.17.4.1) |
C00044491
|
| 23223 | RRP12, KIAA0690 | ribosomal RNA processing 12 homolog (S. cerevisiae) |
C00044491
|
| 51018 | RRP15, KIAA0507 | ribosomal RNA processing 15 homolog (S. cerevisiae) |
C00044491
|
| 88745 | RRP36, C6orf153, dJ20C7.4 | ribosomal RNA processing 36 homolog (S. cerevisiae) |
C00044491
|
| 23212 | RRS1 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
C00044491
|
| 55316 | RSAD1 | radical S-adenosyl methionine domain containing 1 |
C00044491
|
| 91543 | RSAD2, 2510004L01Rik, cig33, cig5, vig1 | radical S-adenosyl methionine domain containing 2 |
C00044491
|
| 222194 | RSBN1L | round spermatid basic protein 1-like |
C00044491
|
| 51187 | RSL24D1, C15orf15, HRP-L30-iso, L30, RLP24, RPL24, RPL24L, TVAS3 | ribosomal L24 domain containing 1 |
C00044491
|
| 83861 | RSPH3, RSHL2, RSP3, dJ111C20.1 | radial spoke 3 homolog (Chlamydomonas) |
C00044491
|
| 81492 | RSPH6A, RSHL1, RSP4, RSP6, RSPH4B | radial spoke head 6 homolog A (Chlamydomonas) |
C00044491
|
| 340419 | RSPO2, CRISTIN2 | R-spondin 2 |
C00044491
|
| 65117 | RSRC2 | arginine/serine-rich coiled-coil 2 |
C00044491
|
| 23168 | RTF1, GTL7, KIAA0252 | Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
C00044491
|
| 6242 | RTKN | rhotekin |
C00044491
|
| 219790 | RTKN2, PLEKHK1, bA531F24.1 | rhotekin 2 |
C00044491
|
| 6252 | RTN1, NSP | reticulon 1 |
C00044491
|
| 64108 | RTP4, IFRG28 | receptor (chemosensory) transporter protein 4 |
C00044491
|
| 25914 | RTTN | rotatin |
C00044491
|
| 55680 | RUFY2, RABIP4R, ZFYVE13 | RUN and FYVE domain containing 2 |
C00044491
|
| 22902 | RUFY3, RIPX, SINGAR1 | RUN and FYVE domain containing 3 |
C00044491
|
| 154661 | RUNDC3B, RPIB9, RPIP9 | RUN domain containing 3B |
C00044491
|
| 284618 | RUSC1-AS1, C1orf104, RP11-21N7.3 | RUSC1 antisense RNA 1 |
C00044491
|
| 8607 | RUVBL1, ECP54, INO80H, NMP238, PONTIN, Pontin52, RVB1, TIH1, TIP49, TIP49A | RuvB-like AAA ATPase 1 (EC:3.6.4.12) |
C00044491
|
| 10856 | RUVBL2, ECP51, INO80J, REPTIN, RVB2, TIH2, TIP48, TIP49B | RuvB-like AAA ATPase 2 (EC:3.6.4.12) |
C00044491
|
| 51289 | RXFP3, GPCR135, RLN3R1, RXFPR3, SALPR | relaxin/insulin-like family peptide receptor 3 |
C00044491
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00044491
|
| 23429 | RYBP, AAP1, DEDAF, YEAF1 | RING1 and YY1 binding protein |
C00044491
|
| 6281 | S100A10, 42C, ANX2L, ANX2LG, CAL1L, CLP11, Ca[1], GP11, P11, p10 | S100 calcium binding protein A10 |
C00044491
|
| 6284 | S100A13 | S100 calcium binding protein A13 |
C00044491
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00044491
|
| 26278 | SACS, ARSACS, DNAJC29 | spastic ataxia of Charlevoix-Saguenay (sacsin) |
C00044491
|
| 6299 | SALL1, HSAL1, Sal-1, TBS, ZNF794 | spalt-like transcription factor 1 |
C00044491
|
| 401474 | SAMD12 | sterile alpha motif domain containing 12 |
C00044491
|
| 23034 | SAMD4A, SAMD4, SMAUG, SMAUG1, SMG, SMGA | sterile alpha motif domain containing 4A |
C00044491
|
| 55095 | SAMD4B, SMGB, hSmaug2 | sterile alpha motif domain containing 4B |
C00044491
|
| 25813 | SAMM50, OMP85, SAM50, TOB55, TRG-3, YNL026W | SAMM50 sorting and assembly machinery component |
C00044491
|
| 79595 | SAP130 | Sin3A-associated protein, 130kDa (EC:3.2.1.3) |
C00044491
|
| 79685 | SAP30L, NS4ATP2 | SAP30-like |
C00044491
|
| 163786 | SASS6, SAS-6, SAS6 | spindle assembly 6 homolog (C. elegans) |
C00044491
|
| 6303 | SAT1, DC21, KFSD, KFSDX, SAT, SSAT, SSAT-1 | spermidine/spermine N1-acetyltransferase 1 (EC:2.3.1.57) |
C00044491
|
| 6304 | SATB1 | SATB homeobox 1 |
C00044491
|
| 23314 | SATB2 | SATB homeobox 2 |
C00044491
|
| 60485 | SAV1, SAV, WW45, WWP4 | salvador homolog 1 (Drosophila) |
C00044491
|
| 55776 | SAYSD1, C6orf64 | SAYSVFN motif domain containing 1 |
C00044491
|
| 6305 | SBF1, CMT4B3, DENND7A, MTMR5 | SET binding factor 1 |
C00044491
|
| 66234 |
C00044491
|
||
| 9169 | SCAF11, CASP11, SFRS2IP, SIP1, SRRP129, SRSF2IP | SR-related CTD-associated factor 11 |
C00044491
|
| 57466 | SCAF4, SFRS15, SRA4 | SR-related CTD-associated factor 4 |
C00044491
|
| 10066 | SCAMP2 | secretory carrier membrane protein 2 |
C00044491
|
| 49855 | SCAPER, ZNF291, Zfp291 | S-phase cyclin A-associated protein in the ER |
C00044491
|
| 286133 | SCARA5, NET33, Tesr | scavenger receptor class A, member 5 (putative) |
C00044491
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00044491
|
| 950 | SCARB2, AMRF, CD36L2, EPM4, HLGP85, LGP85, LIMP-2, LIMPII, SR-BII | scavenger receptor class B, member 2 |
C00044491
|
| 619383 | SCARNA9, Z32, mgU2-19/30 | small Cajal body-specific RNA 9 |
C00044491
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00044491
|
| 23256 | SCFD1, C14orf163, RA410, SLY1, SLY1P, STXBP1L2 | sec1 family domain containing 1 |
C00044491
|
| 152579 | SCFD2, STXBP1L1 | sec1 family domain containing 2 |
C00044491
|
| 7356 | SCGB1A1, CC10, CC16, CCPBP, CCSP, UGB, UP-1, UP1 | secretoglobin, family 1A, member 1 (uteroglobin) |
C00044491
|
| 29970 | SCHIP1, SCHIP-1 | schwannomin interacting protein 1 |
C00044491
|
| 132320 | SCLT1, CAP-1A, CAP1A, hCAP-1A | sodium channel and clathrin linker 1 |
C00044491
|
| 51540 | SCLY, SCL, hSCL | selenocysteine lyase (EC:4.4.1.16) |
C00044491
|
| 10389 | SCML2 | sex comb on midleg-like 2 (Drosophila) |
C00044491
|
| 6324 | SCN1B, ATFB13, BRGDA5, GEFSP1 | sodium channel, voltage-gated, type I, beta subunit |
C00044491
|
| 6337 | SCNN1A, BESC2, ENaCa, ENaCalpha, SCNEA, SCNN1 | sodium channel, non-voltage-gated 1 alpha subunit |
C00044491
|
| 6342 | SCP2, NLTP, NSL-TP, SCP-2, SCP-CHI, SCP-X, SCPX | sterol carrier protein 2 (EC:2.3.1.176) |
C00044491
|
| 59342 | SCPEP1, HSCP1, RISC | serine carboxypeptidase 1 |
C00044491
|
| 23513 | SCRIB, CRIB1, SCRB1, SCRIB1, Vartul | scribbled planar cell polarity protein |
C00044491
|
| 79634 | SCRN3, SES3 | secernin 3 |
C00044491
|
| 6343 | SCT | secretin |
C00044491
|
| 57147 | SCYL3, PACE-1, PACE1 | SCY1-like 3 (S. cerevisiae) |
C00044491
|
| 55153 | SDAD1 | SDA1 domain containing 1 |
C00044491
|
| 6382 | SDC1, CD138, SDC, SYND1, syndecan | syndecan 1 |
C00044491
|
| 6385 | SDC4, SYND4 | syndecan 4 |
C00044491
|
| 163859 | SDE2, C1orf55, RP4-671D7.1, dJ671D7.1 | SDE2 telomere maintenance homolog (S. pombe) |
C00044491
|
| 6388 | SDF2 | stromal cell-derived factor 2 |
C00044491
|
| 113675 | SDSL, SDH_2, SDS-RS1, TDH | serine dehydratase-like (EC:4.3.1.17 4.3.1.19) |
C00044491
|
| 90701 | SEC11C, SEC11L3, SPC21, SPCS4C | SEC11 homolog C (S. cerevisiae) (EC:3.4.21.89) |
C00044491
|
| 6396 | SEC13, D3S1231E, SEC13L1, SEC13R, npp-20 | SEC13 homolog (S. cerevisiae) |
C00044491
|
| 284904 | SEC14L4, TAP3 | SEC14-like 4 (S. cerevisiae) |
C00044491
|
| 9919 | SEC16A, KIAA0310, RP11-413M3.10, SEC16L, p250 | SEC16 homolog A (S. cerevisiae) (EC:3.1.3.36) |
C00044491
|
| 89866 | SEC16B, LZTR2, PGPR-p117, RGPR, SEC16S | SEC16 homolog B (S. cerevisiae) |
C00044491
|
| 26984 | SEC22A, SEC22L2 | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
C00044491
|
| 9117 | SEC22C, SEC22L3 | SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
C00044491
|
| 10484 | SEC23A, CLSD | Sec23 homolog A (S. cerevisiae) |
C00044491
|
| 11196 | SEC23IP, P125, P125A | SEC23 interacting protein |
C00044491
|
| 10802 | SEC24A | SEC24 family member A |
C00044491
|
| 9632 | SEC24C | SEC24 family member C |
C00044491
|
| 29927 | SEC61A1, HSEC61, SEC61, SEC61A | Sec61 alpha 1 subunit (S. cerevisiae) |
C00044491
|
| 55176 | SEC61A2 | Sec61 alpha 2 subunit (S. cerevisiae) |
C00044491
|
| 7095 | SEC62, Dtrp1, HTP1, TLOC1, TP-1 | SEC62 homolog (S. cerevisiae) |
C00044491
|
| 79048 | SECISBP2, SBP2 | SECIS binding protein 2 |
C00044491
|
| 81929 | SEH1L, SEC13L, SEH1A, SEH1B, Seh1 | SEH1-like (S. cerevisiae) |
C00044491
|
| 7869 | SEMA3B, LUCA-1, SEMA5, SEMAA, SemA, semaV | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
C00044491
|
| 6405 | SEMA3F, SEMA-IV, SEMA4, SEMAK | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
C00044491
|
| 10509 | SEMA4B, SEMAC, SemC | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
C00044491
|
| 10505 | SEMA4F, M-SEMA, PRO2353, S4F, SEMAM, SEMAW, m-Sema-M | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
C00044491
|
| 57556 | SEMA6A, HT018, SEMA, SEMA6A1, SEMAQ, VIA | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
C00044491
|
| 59343 | SENP2, AXAM2, SMT3IP2 | SUMO1/sentrin/SMT3 specific peptidase 2 (EC:3.4.22.68) |
C00044491
|
| 26054 | SENP6, SSP1, SUSP1 | SUMO1/sentrin specific peptidase 6 (EC:3.4.22.68) |
C00044491
|
| 57190 | SEPN1, CFTD, MDRS1, RSMD1, RSS, SELN | selenoprotein N, 1 |
C00044491
|
| 51091 | SEPSECS, LP, PCH2D, SLA, SLA/LP | Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (EC:2.9.1.2) |
C00044491
|
| 346288 | SEPT14 | septin 14 |
C00044491
|
| 26135 | SERBP1, CHD3IP, HABP4L, PAI-RBP1, PAIRBP1 | SERPINE1 mRNA binding protein 1 |
C00044491
|
| 26297 | SERGEF, DELGEF, Gnefr | secretion regulating guanine nucleotide exchange factor |
C00044491
|
| 10955 | SERINC3, AIGP1, DIFF33, SBBI99, TDE, TDE1, TMS-1 | serine incorporator 3 |
C00044491
|
| 5265 | SERPINA1, A1A, A1AT, AAT, PI, PI1, PRO2275, alpha1AT | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
C00044491
|
| 51156 | SERPINA10, PZI, ZPI | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
C00044491
|
| 12 | SERPINA3, AACT, ACT, GIG25 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
C00044491
|
| 866 | SERPINA6, CBG | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
C00044491
|
| 5272 | SERPINB9, CAP-3, CAP3, PI-9, PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
C00044491
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00044491
|
| 871 | SERPINH1, AsTP3, CBP1, CBP2, HSP47, OI10, PPROM, RA-A47, SERPINH2, gp46 | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
C00044491
|
| 5274 | SERPINI1, PI12, neuroserpin | serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
C00044491
|
| 29950 | SERTAD1, SEI1, TRIP-Br1 | SERTA domain containing 1 |
C00044491
|
| 9792 | SERTAD2, Sei-2, TRIP-Br2 | SERTA domain containing 2 |
C00044491
|
| 29946 | SERTAD3, RBT1 | SERTA domain containing 3 |
C00044491
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00044491
|
| 83667 | SESN2, HI95, SES2, SEST2 | sestrin 2 |
C00044491
|
| 143686 | SESN3, SEST3 | sestrin 3 |
C00044491
|
| 80854 | SETD7, KMT7, SET7, SET7/9, SET9 | SET domain containing (lysine methyltransferase) 7 (EC:2.1.1.43) |
C00044491
|
| 10291 | SF3A1, PRP21, PRPF21, SAP114, SF3A120 | splicing factor 3a, subunit 1, 120kDa |
C00044491
|
| 23451 | SF3B1, Hsh155, MDS, PRP10, PRPF10, SAP155, SF3b155 | splicing factor 3b, subunit 1, 155kDa |
C00044491
|
| 23450 | SF3B3, RSE1, SAP130, SF3b130, STAF130 | splicing factor 3b, subunit 3, 130kDa |
C00044491
|
| 2810 | SFN, YWHAS | stratifin |
C00044491
|
| 6421 | SFPQ, POMP100, PSF | splicing factor proline/glutamine-rich |
C00044491
|
| 94081 | SFXN1 | sideroflexin 1 |
C00044491
|
| 119559 | SFXN4, BCRM1 | sideroflexin 4 |
C00044491
|
| 94097 | SFXN5, BBG-TCC | sideroflexin 5 |
C00044491
|
| 6443 | SGCB, A3b, LGMD2E, SGC | sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
C00044491
|
| 10110 | SGK2, H-SGK2, dJ138B7.2 | serum/glucocorticoid regulated kinase 2 (EC:2.7.11.1) |
C00044491
|
| 166929 | SGMS2, SMS2 | sphingomyelin synthase 2 (EC:2.7.8.27) |
C00044491
|
| 151648 | SGOL1, NY-BR-85, SGO, Sgo1 | shugoshin-like 1 (S. pombe) |
C00044491
|
| 151246 | SGOL2, SGO2, TRIPIN | shugoshin-like 2 (S. pombe) |
C00044491
|
| 8879 | SGPL1, S1PL, SPL | sphingosine-1-phosphate lyase 1 (EC:4.1.2.27) |
C00044491
|
| 6449 | SGTA, SGT, alphaSGT, hSGT | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
C00044491
|
| 10044 | SH2D3C, CHAT, NSP3, PRO34088, SHEP1 | SH2 domain containing 3C |
C00044491
|
| 63898 | SH2D4A, PPP1R38, SH2A | SH2 domain containing 4A |
C00044491
|
| 284948 | SH2D6 | SH2 domain containing 6 |
C00044491
|
| 9467 | SH3BP5, SAB, SH3BP-5 | SH3-domain binding protein 5 (BTK-associated) |
C00044491
|
| 152503 | SH3D19, EBP, EVE1, Kryn, SH3P19 | SH3 domain containing 19 |
C00044491
|
| 56904 | SH3GLB2, PP6569, PP9455, RRIG1 | SH3-domain GRB2-like endophilin B2 |
C00044491
|
| 285590 | SH3PXD2B, FAD49, FTHS, HOFI, KIAA1295, TKS4, TSK4 | SH3 and PX domains 2B |
C00044491
|
| 50944 | SHANK1, SPANK-1, SSTRIP, synamon | SH3 and multiple ankyrin repeat domains 1 |
C00044491
|
| 6464 | SHC1, SHC, SHCA | SHC (Src homology 2 domain containing) transforming protein 1 |
C00044491
|
| 53358 | SHC3, N-Shc, NSHC, RAI, SHCC | SHC (Src homology 2 domain containing) transforming protein 3 |
C00044491
|
| 79801 | SHCBP1, PAL | SHC SH2-domain binding protein 1 |
C00044491
|
| 56961 | SHD | Src homology 2 domain containing transforming protein D |
C00044491
|
| 6469 | SHH, HHG1, HLP3, HPE3, MCOPCB5, SMMCI, TPT, TPTPS | sonic hedgehog |
C00044491
|
| 51246 | SHISA5, SCOTIN | shisa family member 5 |
C00044491
|
| 92799 | SHKBP1, Sb1 | SH3KBP1 binding protein 1 |
C00044491
|
| 6470 | SHMT1, CSHMT, SHMT | serine hydroxymethyltransferase 1 (soluble) (EC:2.1.2.1) |
C00044491
|
| 6472 | SHMT2, GLYA, SHMT | serine hydroxymethyltransferase 2 (mitochondrial) (EC:2.1.2.1) |
C00044491
|
| 57619 | SHROOM3, APXL3, SHRM, ShrmL | shroom family member 3 |
C00044491
|
| 54414 | SIAE, AIS6, CSE-C, CSEC, LSE, YSG2 | sialic acid acetylesterase (EC:3.1.1.53) |
C00044491
|
| 8778 | SIGLEC5, CD170, CD33L2, OB-BP2, OBBP2, SIGLEC-5 | sialic acid binding Ig-like lectin 5 |
C00044491
|
| 150094 | SIK1, MSK, SIK, SNF1LK | salt-inducible kinase 1 (EC:2.7.11.1) |
C00044491
|
| 23235 | SIK2, LOH11CR1I, QIK, SNF1LK2 | salt-inducible kinase 2 (EC:2.7.11.1) |
C00044491
|
| 375484 | SIMC1, C5orf25, OOMA1, PLEIAD | SUMO-interacting motifs containing 1 |
C00044491
|
| 140885 | SIRPA, BIT, CD172A, MFR, MYD-1, P84, PTPNS1, SHPS1, SIRP | signal-regulatory protein alpha (EC:3.1.3.48) |
C00044491
|
| 23411 | SIRT1, SIR2L1 | sirtuin 1 |
C00044491
|
| 4990 | SIX6, MCOPCT2, OPTX2, Six9 | SIX homeobox 6 |
C00044491
|
| 220134 | SKA1, C18orf24 | spindle and kinetochore associated complex subunit 1 |
C00044491
|
| 348235 | SKA2, FAM33A | spindle and kinetochore associated complex subunit 2 |
C00044491
|
| 221150 | SKA3, C13orf3, RAMA1 | spindle and kinetochore associated complex subunit 3 |
C00044491
|
| 8631 | SKAP1, SCAP1, SKAP55 | src kinase associated phosphoprotein 1 |
C00044491
|
| 23517 | SKIV2L2, Dob1, KIAA0052, Mtr4, fSAP118 | superkiller viralicidic activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13) |
C00044491
|
| 6500 | SKP1, EMC19, OCP-II, OCP2, SKP1A, TCEB1L, p19A | S-phase kinase-associated protein 1 (EC:3.1.1.3) |
C00044491
|
| 6502 | SKP2, FBL1, FBXL1, FLB1, p45 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
C00044491
|
| 6503 | SLA, SLA1, SLAP | Src-like-adaptor |
C00044491
|
| 7884 | SLBP, HBP | stem-loop binding protein |
C00044491
|
| 4891 | SLC11A2, DCT1, DMT1, NRAMP2 | solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
C00044491
|
| 6558 | SLC12A2, BSC, BSC2, NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
C00044491
|
| 6561 | SLC13A1, NAS1, NaSi-1 | solute carrier family 13 (sodium/sulfate symporter), member 1 |
C00044491
|
| 284111 | SLC13A5, NACT | solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
C00044491
|
| 121260 | SLC15A4, PHT1, PTR4 | solute carrier family 15 (oligopeptide transporter), member 4 |
C00044491
|
| 9120 | SLC16A6, MCT6, MCT7 | solute carrier family 16, member 6 |
C00044491
|
| 9194 | SLC16A7, MCT2 | solute carrier family 16 (monocarboxylate transporter), member 7 |
C00044491
|
| 10246 | SLC17A2, NPT3 | solute carrier family 17, member 2 |
C00044491
|
| 63910 | SLC17A9, C20orf59, VNUT | solute carrier family 17 (vesicular nucleotide transporter), member 9 |
C00044491
|
| 116843 | SLC18B1, C6orf192, dJ55C23.6 | solute carrier family 18, subfamily B, member 1 |
C00044491
|
| 10560 | SLC19A2, TC1, THMD1, THT1, THTR1, TRMA | solute carrier family 19 (thiamine transporter), member 2 |
C00044491
|
| 6574 | SLC20A1, GLVR1, Glvr-1, PIT1, PiT-1 | solute carrier family 20 (phosphate transporter), member 1 |
C00044491
|
| 55867 | SLC22A11, OAT4, hOAT4 | solute carrier family 22 (organic anion/urate transporter), member 11 |
C00044491
|
| 9389 | SLC22A14, OCTL2, OCTL4, ORCTL4 | solute carrier family 22, member 14 |
C00044491
|
| 63027 | SLC22A23, C6orf85 | solute carrier family 22, member 23 |
C00044491
|
| 6581 | SLC22A3, EMT, EMTH, OCT3 | solute carrier family 22 (organic cation transporter), member 3 |
C00044491
|
| 9356 | SLC22A6, HOAT1, OAT1, PAHT, ROAT1 | solute carrier family 22 (organic anion transporter), member 6 |
C00044491
|
| 10864 | SLC22A7, NLT, OAT2 | solute carrier family 22 (organic anion transporter), member 7 |
C00044491
|
| 114571 | SLC22A9, HOAT4, OAT4, OAT7, UST3H, ust3 | solute carrier family 22 (organic anion transporter), member 9 |
C00044491
|
| 9962 | SLC23A2, NBTL1, SLC23A1, SVCT2, YSPL2 | solute carrier family 23 (ascorbic acid transporter), member 2 |
C00044491
|
| 10165 | SLC25A13, ARALAR2, CITRIN, CTLN2 | solute carrier family 25 (aspartate/glutamate carrier), member 13 |
C00044491
|
| 8034 | SLC25A16, D10S105E, GDA, GDC, HGT.1, ML7, hML7 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
C00044491
|
| 114789 | SLC25A25, MCSC, PCSCL, RP11-395P17.4, SCAMC-2 | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
C00044491
|
| 9481 | SLC25A27, UCP4 | solute carrier family 25, member 27 |
C00044491
|
| 84275 | SLC25A33, BMSC-MCP, PNC1 | solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
C00044491
|
| 51312 | SLC25A37, MFRN, MFRN1, MSC, MSCP, PRO1278, PRO1584, PRO2217 | solute carrier family 25 (mitochondrial iron transporter), member 37 |
C00044491
|
| 283130 | SLC25A45 | solute carrier family 25, member 45 |
C00044491
|
| 91137 | SLC25A46 | solute carrier family 25, member 46 |
C00044491
|
| 283600 | SLC25A47, C14orf68, HDMCP | solute carrier family 25, member 47 |
C00044491
|
| 65010 | SLC26A6 | solute carrier family 26 (anion exchanger), member 6 |
C00044491
|
| 11000 | SLC27A3, ACSVL3, FATP3, VLCS-3 | solute carrier family 27 (fatty acid transporter), member 3 (EC:6.2.1.-) |
C00044491
|
| 10999 | SLC27A4, ACSVL4, FATP4, IPS | solute carrier family 27 (fatty acid transporter), member 4 (EC:6.2.1.-) |
C00044491
|
| 9154 | SLC28A1, CNT1, HCNT1 | solute carrier family 28 (concentrative nucleoside transporter), member 1 |
C00044491
|
| 2030 | SLC29A1, ENT1 | solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
C00044491
|
| 6514 | SLC2A2, GLUT2 | solute carrier family 2 (facilitated glucose transporter), member 2 |
C00044491
|
| 6515 | SLC2A3, GLUT3 | solute carrier family 2 (facilitated glucose transporter), member 3 |
C00044491
|
| 100128062 |
C00044491
|
||
| 29988 | SLC2A8, GLUT8, GLUTX1 | solute carrier family 2 (facilitated glucose transporter), member 8 |
C00044491
|
| 7779 | SLC30A1, ZNT1, ZRC1 | solute carrier family 30 (zinc transporter), member 1 |
C00044491
|
| 55676 | SLC30A6, MST103, MSTP103, ZNT6 | solute carrier family 30 (zinc transporter), member 6 |
C00044491
|
| 148867 | SLC30A7, ZNT7, ZnT-7, ZnTL2 | solute carrier family 30 (zinc transporter), member 7 |
C00044491
|
| 1317 | SLC31A1, COPT1, CTR1 | solute carrier family 31 (copper transporter), member 1 |
C00044491
|
| 9197 | SLC33A1, ACATN, AT-1, AT1, CCHLND, SPG42 | solute carrier family 33 (acetyl-CoA transporter), member 1 |
C00044491
|
| 23443 | SLC35A3 | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
C00044491
|
| 347734 | SLC35B2, PAPST1, SLL, UGTrel4 | solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
C00044491
|
| 84912 | SLC35B4, YEA, YEA4 | solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
C00044491
|
| 55343 | SLC35C1, CDG2C, FUCT1 | solute carrier family 35 (GDP-fucose transporter), member C1 |
C00044491
|
| 23169 | SLC35D1, UGTREL7 | solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
C00044491
|
| 11046 | SLC35D2, HFRC1, SQV7L, UGTrel8, hfrc | solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
C00044491
|
| 79939 | SLC35E1 | solute carrier family 35, member E1 |
C00044491
|
| 728661 | SLC35E2B, SLC35E2 | solute carrier family 35, member E2B |
C00044491
|
| 55508 | SLC35E3, BLOV1 | solute carrier family 35, member E3 |
C00044491
|
| 148641 | SLC35F3 | solute carrier family 35, member F3 |
C00044491
|
| 80255 | SLC35F5 | solute carrier family 35, member F5 |
C00044491
|
| 54978 | SLC35F6, ANT2BP, C2orf18, TANGO9 | solute carrier family 35, member F6 |
C00044491
|
| 206358 | SLC36A1, Dct1, LYAAT1, PAT1, TRAMD3 | solute carrier family 36 (proton/amino acid symporter), member 1 |
C00044491
|
| 120103 | SLC36A4, PAT4 | solute carrier family 36 (proton/amino acid symporter), member 4 |
C00044491
|
| 81539 | SLC38A1, ATA1, NAT2, SAT1, SNAT1 | solute carrier family 38, member 1 |
C00044491
|
| 124565 | SLC38A10 | solute carrier family 38, member 10 |
C00044491
|
| 151258 | SLC38A11, AVT2 | solute carrier family 38, member 11 |
C00044491
|
| 55089 | SLC38A4, ATA3, NAT3, PAAT | solute carrier family 38, member 4 |
C00044491
|
| 92745 | SLC38A5, JM24, SN2, SNAT5, pp7194 | solute carrier family 38, member 5 |
C00044491
|
| 153129 | SLC38A9 | solute carrier family 38, member 9 |
C00044491
|
| 23516 | SLC39A14, LZT-Hs4, NET34, ZIP14, cig19 | solute carrier family 39 (zinc transporter), member 14 |
C00044491
|
| 283375 | SLC39A5, LZT-Hs7, ZIP5 | solute carrier family 39 (zinc transporter), member 5 |
C00044491
|
| 6519 | SLC3A1, ATR1, CSNU1, D2H, NBAT, RBAT | solute carrier family 3 (amino acid transporter heavy chain), member 1 |
C00044491
|
| 30061 | SLC40A1, FPN1, HFE4, IREG1, MST079, MTP1, SLC11A3 | solute carrier family 40 (iron-regulated transporter), member 1 |
C00044491
|
| 254428 | SLC41A1, MgtE | solute carrier family 41 (magnesium transporter), member 1 |
C00044491
|
| 84102 | SLC41A2, SLC41A1-L1 | solute carrier family 41 (magnesium transporter), member 2 |
C00044491
|
| 8501 | SLC43A1, LAT3, PB39, POV1, R00504 | solute carrier family 43 (amino acid system L transporter), member 1 |
C00044491
|
| 124935 | SLC43A2, LAT4 | solute carrier family 43 (amino acid system L transporter), member 2 |
C00044491
|
| 29015 | SLC43A3, EEG1, FOAP-13, PRO1659, SEEEG-1 | solute carrier family 43, member 3 |
C00044491
|
| 85414 | SLC45A3, IPCA-2, IPCA-6, IPCA-8, IPCA6, PCANAP2, PCANAP6, PCANAP8, PRST | solute carrier family 45, member 3 |
C00044491
|
| 55652 | SLC48A1, HRG-1, HRG1, hHRG-1 | solute carrier family 48 (heme transporter), member 1 |
C00044491
|
| 57282 | SLC4A10, NBCn2, NCBE | solute carrier family 4, sodium bicarbonate transporter, member 10 |
C00044491
|
| 22950 | SLC4A1AP | solute carrier family 4 (anion exchanger), member 1, adaptor protein |
C00044491
|
| 8671 | SLC4A4, HNBC1, KNBC, NBC1, NBC2, NBCe1-A, SLC4A5, hhNMC, pNBC | solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
C00044491
|
| 9498 | SLC4A8, NBC3, NDCBE | solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
C00044491
|
| 6526 | SLC5A3, SMIT, SMIT1, SMIT2 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
C00044491
|
| 200010 | SLC5A9, SGLT4 | solute carrier family 5 (sodium/sugar cotransporter), member 9 |
C00044491
|
| 6535 | SLC6A8, CCDS1, CRT, CRTR, CT1 | solute carrier family 6 (neurotransmitter transporter), member 8 |
C00044491
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00044491
|
| 6542 | SLC7A2, ATRC2, CAT2, HCAT2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
C00044491
|
| 8140 | SLC7A5, 4F2LC, CD98, D16S469E, E16, LAT1, MPE16, hLAT1 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
C00044491
|
| 84138 | SLC7A6OS | solute carrier family 7, member 6 opposite strand |
C00044491
|
| 9056 | SLC7A7, LAT3, LPI, MOP-2, Y+LAT1, y+LAT-1 | solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
C00044491
|
| 6543 | SLC8A2, NCX2 | solute carrier family 8 (sodium/calcium exchanger), member 2 |
C00044491
|
| 6548 | SLC9A1, APNH, NHE-1, NHE1 | solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
C00044491
|
| 9351 | SLC9A3R2, E3KARP, NHE3RF2, NHERF-2, NHERF2, OCTS2, SIP-1, SIP1, TKA-1 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
C00044491
|
| 84679 | SLC9A7, NHE-7, NHE7, SLC9A6 | solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
C00044491
|
| 162394 | SLFN5 | schlafen family member 5 |
C00044491
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00044491
|
| 4092 | SMAD7, CRCS3, MADH7, MADH8 | SMAD family member 7 |
C00044491
|
| 57228 | SMAGP, hSMAGP | small cell adhesion glycoprotein |
C00044491
|
| 6594 | SMARCA1, ISWI, NURF140, SNF2L, SNF2L1, SNF2LB, SNF2LT, SWI, SWI2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
C00044491
|
| 6595 | SMARCA2, BAF190, BRM, NCBRS, SNF2, SNF2L2, SNF2LA, SWI2, Sth1p, hBRM, hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
C00044491
|
| 6597 | SMARCA4, BAF190, BAF190A, BRG1, MRD16, RTPS2, SNF2, SNF2L4, SNF2LB, SWI2, hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
C00044491
|
| 8467 | SMARCA5, ISWI, SNF2H, WCRF135, hISWI, hSNF2H | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
C00044491
|
| 56916 | SMARCAD1, ADERM, ETL1, HEL1 | SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 (EC:3.6.4.12) |
C00044491
|
| 50485 | SMARCAL1, HARP, HHARP | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
C00044491
|
| 6605 | SMARCE1, BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
C00044491
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00044491
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00044491
|
| 9126 | SMC3, BAM, BMH, CDLS3, CSPG6, HCAP, SMC3L1 | structural maintenance of chromosomes 3 |
C00044491
|
| 10051 | SMC4, CAP-C, CAPC, SMC-4, SMC4L1, hCAP-C | structural maintenance of chromosomes 4 |
C00044491
|
| 79677 | SMC6, SMC-6, SMC6L1, hSMC6 | structural maintenance of chromosomes 6 |
C00044491
|
| 55671 | SMEK1, FLFL1, KIAA2010, PP4R3A, smk-1, smk1 | SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
C00044491
|
| 57223 | SMEK2, FLFL2, PP4R3B, PSY2, smk1 | SMEK homolog 2, suppressor of mek1 (Dictyostelium) |
C00044491
|
| 201895 | SMIM14, C4orf34 | small integral membrane protein 14 |
C00044491
|
| 440957 | SMIM4, C3orf78 | small integral membrane protein 4 |
C00044491
|
| 79086 | SMIM7, C19orf42 | small integral membrane protein 7 |
C00044491
|
| 54498 | SMOX, C20orf16, PAO, PAO-1, PAOH1, SMO | spermine oxidase (EC:1.5.3.16) |
C00044491
|
| 6609 | SMPD1, ASM, ASMASE, NPD | sphingomyelin phosphodiesterase 1, acid lysosomal (EC:3.1.4.12) |
C00044491
|
| 10924 | SMPDL3A, ASM3A, ASML3a, yR36GH4.1 | sphingomyelin phosphodiesterase, acid-like 3A |
C00044491
|
| 6525 | SMTN | smoothelin |
C00044491
|
| 56950 | SMYD2, HSKM-B, KMT3C, ZMYND14 | SET and MYND domain containing 2 (EC:2.1.1.43) |
C00044491
|
| 114826 | SMYD4, ZMYND21 | SET and MYND domain containing 4 |
C00044491
|
| 6591 | SNAI2, SLUG, SLUGH1, SNAIL2, WS2D | snail family zinc finger 2 |
C00044491
|
| 8773 | SNAP23, HsT17016, SNAP-23, SNAP23A, SNAP23B | synaptosomal-associated protein, 23kDa |
C00044491
|
| 9342 | SNAP29, CEDNIK, SNAP-29 | synaptosomal-associated protein, 29kDa |
C00044491
|
| 23642 | SNHG1, LINC00057, NCRNA00057, U22HG, UHG | small nucleolar RNA host gene 1 (non-protein coding) |
C00044491
|
| 85028 | SNHG12, C1orf79, LINC00100, NCRNA00100, RP4-669K10.5 | small nucleolar RNA host gene 12 (non-protein coding) |
C00044491
|
| 100093630 | SNHG8, LINC00060, NCRNA00060 | small nucleolar RNA host gene 8 (non-protein coding) |
C00044491
|
| 8303 | SNN | stannin |
C00044491
|
| 677811 | SNORA28, ACA28 | small nucleolar RNA, H/ACA box 28 |
C00044491
|
| 677816 | SNORA35, HBI-36 | small nucleolar RNA, H/ACA box 35 |
C00044491
|
| 677830 | SNORA50, ACA50 | small nucleolar RNA, H/ACA box 50 |
C00044491
|
| 26821 | SNORA74A, RNU19, U19 | small nucleolar RNA, H/ACA box 74A |
C00044491
|
| 654321 | SNORA75, U23 | small nucleolar RNA, H/ACA box 75 |
C00044491
|
| 100033430 | SNORD116-18, HBII-85-18 | small nucleolar RNA, C/D box 116-18 |
C00044491
|
| 26814 | SNORD36B, RNU36B, U36b | small nucleolar RNA, C/D box 36B |
C00044491
|
| 26806 | SNORD44, RNU44, U44 | small nucleolar RNA, C/D box 44 |
C00044491
|
| 26799 | SNORD50A, RNU50, U50 | small nucleolar RNA, C/D box 50A |
C00044491
|
| 26791 | SNORD58A, RNU58A, U58a | small nucleolar RNA, C/D box 58A |
C00044491
|
| 619498 | SNORD74, U74, Z18 | small nucleolar RNA, C/D box 74 |
C00044491
|
| 26770 | SNORD79, RNU103, U79, Z22 | small nucleolar RNA, C/D box 79 |
C00044491
|
| 25826 | SNORD82, RNU82, U82, Z25 | small nucleolar RNA, C/D box 82 |
C00044491
|
| 692205 | SNORD89, HBII-289 | small nucleolar RNA, C/D box 89 |
C00044491
|
| 11066 | SNRNP35, HM-1, U1SNRNPBP | small nuclear ribonucleoprotein 35kDa (U11/U12) |
C00044491
|
| 29887 | SNX10, OPTB8 | sorting nexin 10 |
C00044491
|
| 57231 | SNX14, RGS-PX2 | sorting nexin 14 |
C00044491
|
| 112574 | SNX18, SH3PX2, SH3PXD3B, SNAG1 | sorting nexin 18 |
C00044491
|
| 399979 | SNX19, CHET8 | sorting nexin 19 |
C00044491
|
| 81609 | SNX27, MRT1, MY014 | sorting nexin family member 27 |
C00044491
|
| 6646 | SOAT1, ACACT, ACAT, ACAT-1, ACAT1, SOAT, STAT | sterol O-acyltransferase 1 (EC:2.3.1.26) |
C00044491
|
| 8835 | SOCS2, CIS2, Cish2, SOCS-2, SSI-2, SSI2, STATI2 | suppressor of cytokine signaling 2 |
C00044491
|
| 122809 | SOCS4, SOCS7 | suppressor of cytokine signaling 4 |
C00044491
|
| 9655 | SOCS5, CIS6, CISH6, Cish5, SOCS-5 | suppressor of cytokine signaling 5 |
C00044491
|
| 9306 | SOCS6, CIS-4, CIS4, HSPC060, SOCS-4, SOCS-6, SOCS4, SSI4, STAI4, STATI4 | suppressor of cytokine signaling 6 |
C00044491
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00044491
|
| 140710 | SOGA1, C20orf117, KIAA0889, SOGA | suppressor of glucose, autophagy associated 1 |
C00044491
|
| 23255 | SOGA2, CCDC165, KIAA0802 | SOGA family member 2 |
C00044491
|
| 8470 | SORBS2, ARGBP2, PRO0618 | sorbin and SH3 domain containing 2 |
C00044491
|
| 6653 | SORL1, C11orf32, LR11, LRP9, SORLA, SorLA-1, gp250 | sortilin-related receptor, L(DLR class) A repeats containing |
C00044491
|
| 65124 | SOWAHC, ANKRD57, C2orf26 | sosondowah ankyrin repeat domain family member C |
C00044491
|
| 64321 | SOX17, VUR3 | SRY (sex determining region Y)-box 17 |
C00044491
|
| 6672 | SP100, lysp100b | SP100 nuclear antigen |
C00044491
|
| 3431 | SP110, IFI41, IFI75, IPR1, VODI | SP110 nuclear body protein |
C00044491
|
| 93349 | SP140L | SP140 nuclear body protein-like |
C00044491
|
| 6671 | SP4, HF1B, SPR-1 | Sp4 transcription factor |
C00044491
|
| 6674 | SPAG1, CILD28, HSD-3.8, SP75, TPIS | sperm associated antigen 1 |
C00044491
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00044491
|
| 9043 | SPAG9, CT89, HLC-6, HLC4, JIP-4, JIP4, JLP, PHET, PIG6 | sperm associated antigen 9 |
C00044491
|
| 139067 | SPANXN3, CT11.8, SPANX-N3 | SPANX family, member N3 |
C00044491
|
| 6683 | SPAST, ADPSP, FSP2, SPG4 | spastin (EC:3.6.4.3) |
C00044491
|
| 132671 | SPATA18, Mieap, SPETEX1 | spermatogenesis associated 18 |
C00044491
|
| 9825 | SPATA2, PD1, tamo | spermatogenesis associated 2 |
C00044491
|
| 124044 | SPATA2L, C16orf76, tamo | spermatogenesis associated 2-like |
C00044491
|
| 166378 | SPATA5, AFG2, SPAF | spermatogenesis associated 5 |
C00044491
|
| 55812 | SPATA7, HSD-3.1, HSD3, LCA3 | spermatogenesis associated 7 |
C00044491
|
| 26010 | SPATS2L, DNAPTP6, SGNP | spermatogenesis associated, serine-rich 2-like |
C00044491
|
| 147841 | SPC24, SPBC24 | SPC24, NDC80 kinetochore complex component |
C00044491
|
| 387778 | SPDYC, RINGOC, Ringo2 | speedy/RINGO cell cycle regulator family member C |
C00044491
|
| 23384 | SPECC1L, CYTSA, OBLFC1 | sperm antigen with calponin homology and coiled-coil domains 1-like |
C00044491
|
| 80208 | SPG11, KIAA1840 | spastic paraplegia 11 (autosomal recessive) |
C00044491
|
| 8877 | SPHK1, SPHK | sphingosine kinase 1 (EC:2.7.1.91) |
C00044491
|
| 152185 | SPICE1, CCDC52, SPICE | spindle and centriole associated protein 1 |
C00044491
|
| 23514 | SPIDR, KIAA0146 | scaffolding protein involved in DNA repair |
C00044491
|
| 6693 | SPN, CD43, GALGP, GPL115, LSN | sialophorin |
C00044491
|
| 90853 | SPOCD1 | SPOC domain containing 1 |
C00044491
|
| 10417 | SPON2, DIL-1, DIL1, M-SPONDIN, MINDIN | spondin 2, extracellular matrix protein |
C00044491
|
| 200734 | SPRED2, Spred-2 | sprouty-related, EVH1 domain containing 2 |
C00044491
|
| 83932 | SPRTN, C1orf124, DDDL1880, DVC1, PRO4323, Spartan, dJ876B10.3 | SprT-like N-terminal domain |
C00044491
|
| 81848 | SPRY4, HH17 | sprouty homolog 4 (Drosophila) |
C00044491
|
| 80176 | SPSB1, SSB-1, SSB1 | splA/ryanodine receptor domain and SOCS box containing 1 |
C00044491
|
| 6709 | SPTAN1, EIEE5, NEAS, SPTA2 | spectrin, alpha, non-erythrocytic 1 |
C00044491
|
| 6711 | SPTBN1, ELF, SPTB2, betaSpII | spectrin, beta, non-erythrocytic 1 |
C00044491
|
| 9517 | SPTLC2, HSN1C, LCB2, LCB2A, NSAN1C, SPT2, hLCB2a | serine palmitoyltransferase, long chain base subunit 2 (EC:2.3.1.50) |
C00044491
|
| 55304 | SPTLC3, C20orf38, LCB2B, LCB3, SPT3, SPTLC2L, dJ718P11, dJ718P11.1, hLCB2b | serine palmitoyltransferase, long chain base subunit 3 (EC:2.3.1.50) |
C00044491
|
| 6713 | SQLE | squalene epoxidase (EC:1.14.13.132) |
C00044491
|
| 58472 | SQRDL, PRO1975 | sulfide quinone reductase-like (yeast) |
C00044491
|
| 8878 | SQSTM1, A170, OSIL, PDB3, ZIP3, p60, p62, p62B | sequestosome 1 |
C00044491
|
| 80725 | SRCIN1, P140, SNIP | SRC kinase signaling inhibitor 1 |
C00044491
|
| 23380 | SRGAP2, ARHGAP34, FNBP2, SRGAP2A, SRGAP3 | SLIT-ROBO Rho GTPase activating protein 2 |
C00044491
|
| 653464 | SRGAP2C, SRGAP2P1 | SLIT-ROBO Rho GTPase activating protein 2C |
C00044491
|
| 6733 | SRPK2, SFRSK2 | SRSF protein kinase 2 (EC:2.7.11.1) |
C00044491
|
| 27286 | SRPX2, BPP, CBPS, PMGX, RESDX, SRPUL | sushi-repeat containing protein, X-linked 2 |
C00044491
|
| 84530 | SRRM4, KIAA1853, MU-MB-2.76, nSR100 | serine/arginine repetitive matrix 4 |
C00044491
|
| 6426 | SRSF1, ASF, SF2, SF2p33, SFRS1, SRp30a | serine/arginine-rich splicing factor 1 |
C00044491
|
| 6428 | SRSF3, SFRS3, SRp20 | serine/arginine-rich splicing factor 3 |
C00044491
|
| 6430 | SRSF5, HRS, SFRS5, SRP40 | serine/arginine-rich splicing factor 5 |
C00044491
|
| 6432 | SRSF7, 9G8, AAG3, SFRS7 | serine/arginine-rich splicing factor 7 |
C00044491
|
| 10929 | SRSF8, DSM-1, SFRS2B, SRP46 | serine/arginine-rich splicing factor 8 |
C00044491
|
| 140809 | SRXN1, C20orf139, Npn3, SRX, SRX1 | sulfiredoxin 1 (EC:1.8.98.2) |
C00044491
|
| 6741 | SSB, LARP3, La | Sjogren syndrome antigen B (autoantigen La) |
C00044491
|
| 6742 | SSBP1, Mt-SSB, SOSS-B1, SSBP, mtSSB | single-stranded DNA binding protein 1, mitochondrial |
C00044491
|
| 85464 | SSH2, SSH-2, SSH-2L | slingshot protein phosphatase 2 (EC:3.1.3.16 3.1.3.48) |
C00044491
|
| 29101 | SSU72, HSPC182, PNAS-120 | SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) (EC:3.1.3.16) |
C00044491
|
| 6487 | ST3GAL3, EIEE15, MRT12, SIAT6, ST3GALII, ST3GalIII, ST3N | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 (EC:2.4.99.6) |
C00044491
|
| 6764 | ST5, DENND2B, HTS1, p126 | suppression of tumorigenicity 5 |
C00044491
|
| 6480 | ST6GAL1, SIAT1, ST6GalI, ST6N | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (EC:2.4.99.1) |
C00044491
|
| 23166 | STAB1, CLEVER-1, FEEL-1, FELE-1, FEX1, STAB-1 | stabilin 1 |
C00044491
|
| 10274 | STAG1, SA1, SCC3A | stromal antigen 1 |
C00044491
|
| 10735 | STAG2, SA-2, SA2, SCC3B, bA517O1.1 | stromal antigen 2 |
C00044491
|
| 10254 | STAM2, Hbp, STAM2A, STAM2B | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
C00044491
|
| 57559 | STAMBPL1, ALMalpha, AMSH-FP, AMSH-LP, bA399O19.2 | STAM binding protein-like 1 (EC:3.1.2.15) |
C00044491
|
| 55620 | STAP2, BKS | signal transducing adaptor family member 2 |
C00044491
|
| 10809 | STARD10, NY-CO-28, PCTP2, SDCCAG28 | StAR-related lipid transfer (START) domain containing 10 |
C00044491
|
| 80765 | STARD5 | StAR-related lipid transfer (START) domain containing 5 |
C00044491
|
| 57519 | STARD9 | StAR-related lipid transfer (START) domain containing 9 |
C00044491
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00044491
|
| 6773 | STAT2, ISGF-3, P113, STAT113 | signal transducer and activator of transcription 2, 113kDa |
C00044491
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00044491
|
| 6778 | STAT6, D12S1644, IL-4-STAT, STAT6B, STAT6C | signal transducer and activator of transcription 6, interleukin-4 induced |
C00044491
|
| 6781 | STC1, STC | stanniocalcin 1 |
C00044491
|
| 26872 | STEAP1, PRSS24, STEAP | six transmembrane epithelial antigen of the prostate 1 |
C00044491
|
| 6491 | STIL, MCPH7, SIL | SCL/TAL1 interrupting locus |
C00044491
|
| 57620 | STIM2 | stromal interaction molecule 2 |
C00044491
|
| 10963 | STIP1, HOP, IEF-SSP-3521, P60, STI1, STI1L | stress-induced-phosphoprotein 1 |
C00044491
|
| 9263 | STK17A, DRAK1 | serine/threonine kinase 17a (EC:2.7.11.1) |
C00044491
|
| 9262 | STK17B, DRAK2 | serine/threonine kinase 17b (EC:2.7.11.1) |
C00044491
|
| 6788 | STK3, KRS1, MST2 | serine/threonine kinase 3 (EC:2.7.11.1) |
C00044491
|
| 11329 | STK38, NDR, NDR1 | serine/threonine kinase 38 (EC:2.7.11.1) |
C00044491
|
| 6789 | STK4, KRS2, MST1, TIIAC, YSK3 | serine/threonine kinase 4 (EC:2.7.11.1) |
C00044491
|
| 83931 | STK40, SHIK, SgK495 | serine/threonine kinase 40 (EC:2.7.11.1) |
C00044491
|
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00044491
|
| 2040 | STOM, BND7, EPB7, EPB72 | stomatin |
C00044491
|
| 56977 | STOX2 | storkhead box 2 |
C00044491
|
| 11171 | STRAP, MAWD, PT-WD, UNRIP | serine/threonine kinase receptor associated protein |
C00044491
|
| 55342 | STRBP, ILF3L, SPNR, p74 | spermatid perinuclear RNA binding protein |
C00044491
|
| 57464 | STRIP2, FAM40B, FAR11B | striatin interacting protein 2 |
C00044491
|
| 6801 | STRN, SG2NA | striatin, calmodulin binding protein |
C00044491
|
| 29966 | STRN3, SG2NA | striatin, calmodulin binding protein 3 |
C00044491
|
| 412 | STS, ARSC, ARSC1, ASC, ES, SSDD, XLI | steroid sulfatase (microsomal), isozyme S (EC:3.1.6.2) |
C00044491
|
| 3703 | STT3A, ITM1, STT3-A, TMC | STT3A, subunit of the oligosaccharyltransferase complex (catalytic) (EC:2.4.99.18) |
C00044491
|
| 23673 | STX12, STX13, STX14 | syntaxin 12 |
C00044491
|
| 8675 | STX16, SYN16 | syntaxin 16 |
C00044491
|
| 2054 | STX2, EPIM, EPM, STX2A, STX2B, STX2C | syntaxin 2 |
C00044491
|
| 6809 | STX3, STX3A | syntaxin 3 |
C00044491
|
| 6812 | STXBP1, MUNC18-1, NSEC1, P67, RBSEC1, UNC18 | syntaxin binding protein 1 |
C00044491
|
| 6814 | STXBP3, MUNC18-3, MUNC18C, PSP, UNC-18C | syntaxin binding protein 3 |
C00044491
|
| 55959 | SULF2, HSULF-2 | sulfatase 2 |
C00044491
|
| 27233 | SULT1C4, SULT1C, SULT1C2 | sulfotransferase family, cytosolic, 1C, member 4 (EC:2.8.2.-) |
C00044491
|
| 23353 | SUN1, UNC84A | Sad1 and UNC84 domain containing 1 |
C00044491
|
| 8464 | SUPT3H, SPT3, SPT3L | suppressor of Ty 3 homolog (S. cerevisiae) |
C00044491
|
| 9913 | SUPT7L, SPT7L, STAF65, STAF65(gamma), STAF65G, SUPT7H | suppressor of Ty 7 (S. cerevisiae)-like |
C00044491
|
| 64420 | SUSD1 | sushi domain containing 1 |
C00044491
|
| 203328 | SUSD3 | sushi domain containing 3 |
C00044491
|
| 6839 | SUV39H1, H3-K9-HMTase_1, KMT1A, MG44, SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00044491
|
| 51111 | SUV420H1, CGI85, KMT5B | suppressor of variegation 4-20 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00044491
|
| 84787 | SUV420H2, KMT5C | suppressor of variegation 4-20 homolog 2 (Drosophila) (EC:2.1.1.43) |
C00044491
|
| 23512 | SUZ12, CHET9, JJAZ1 | SUZ12 polycomb repressive complex 2 subunit |
C00044491
|
| 6840 | SVIL | supervillin |
C00044491
|
| 23075 | SWAP70, SWAP-70 | SWAP switching B-cell complex 70kDa subunit |
C00044491
|
| 94056 | SYAP1 | synapse associated protein 1 |
C00044491
|
| 6847 | SYCP1, CT8, HOM-TES-14, SCP-1, SCP1 | synaptonemal complex protein 1 |
C00044491
|
| 8189 | SYMPK, SPK, SYM | symplekin |
C00044491
|
| 81493 | SYNC, SYNC1, SYNCOILIN | syncoilin, intermediate filament protein |
C00044491
|
| 10492 | SYNCRIP, GRY-RBP, GRYRBP, HNRNPQ, HNRPQ1, NSAP1, PP68, hnRNP-Q | synaptotagmin binding, cytoplasmic RNA interacting protein |
C00044491
|
| 23345 | SYNE1, 8B, ARCA1, C6orf98, CPG2, EDMD4, MYNE1, Nesp1, SCAR8, dJ45H2.2 | spectrin repeat containing, nuclear envelope 1 |
C00044491
|
| 23224 | SYNE2, EDMD5, NUA, NUANCE, Nesp2, Nesprin-2, SYNE-2, TROPH | spectrin repeat containing, nuclear envelope 2 |
C00044491
|
| 55333 | SYNJ2BP, ARIP2, OMP25 | synaptojanin 2 binding protein |
C00044491
|
| 23336 | SYNM, DMN, SYN | synemin, intermediate filament protein |
C00044491
|
| 284612 | SYPL2, MG29 | synaptophysin-like 2 |
C00044491
|
| 51760 | SYT17 | synaptotagmin XVII |
C00044491
|
| 148281 | SYT6, sytVI | synaptotagmin VI |
C00044491
|
| 9066 | SYT7, IPCA-7, IPCA7, PCANAP7, SYT-VII, SYTVII | synaptotagmin VII |
C00044491
|
| 84958 | SYTL1, JFC1, SLP1 | synaptotagmin-like 1 |
C00044491
|
| 54843 | SYTL2, CHR11SYT, EXO4, SGA72M, SLP2, SLP2A | synaptotagmin-like 2 |
C00044491
|
| 26099 | SZRD1, C1orf144 | SUZ RNA binding domain containing 1 |
C00044491
|
| 6862 | T, TFT | T, brachyury homolog (mouse) |
C00044491
|
| 100505477 |
C00044491
|
||
| 257397 | TAB3, MAP3K7IP3, NAP1 | TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
C00044491
|
| 6863 | TAC1, Hs.2563, NK2, NKNA, NPK, TAC2 | tachykinin, precursor 1 |
C00044491
|
| 10579 | TACC2, AZU-1, ECTACC | transforming, acidic coiled-coil containing protein 2 |
C00044491
|
| 10460 | TACC3, ERIC-1, ERIC1 | transforming, acidic coiled-coil containing protein 3 |
C00044491
|
| 51204 | TACO1, CCDC44 | translational activator of mitochondrially encoded cytochrome c oxidase I |
C00044491
|
| 6882 | TAF11, MGC:15243, TAF2I, TAFII28 | TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
C00044491
|
| 6883 | TAF12, TAF2J, TAFII20 | TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
C00044491
|
| 8148 | TAF15, Npl3, RBP56, TAF2N, TAFII68 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
C00044491
|
| 83860 | TAF3, TAF140, TAFII-140, TAFII140 | TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
C00044491
|
| 6877 | TAF5, TAF2D, TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
C00044491
|
| 6878 | TAF6, MGC:8964, TAF(II)70, TAF(II)80, TAF2E, TAFII-70, TAFII-80, TAFII70, TAFII80, TAFII85 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
C00044491
|
| 6879 | TAF7, TAF2F, TAFII55 | TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
C00044491
|
| 129685 | TAF8, 43, II, TAF, TAFII-43, TAFII43, TBN | TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
C00044491
|
| 6876 | TAGLN, SM22, SMCC, TAGLN1, WS3-10 | transgelin |
C00044491
|
| 8407 | TAGLN2, HA1756 | transgelin 2 |
C00044491
|
| 85461 | TANC1, ROLSB, TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
C00044491
|
| 128989 | TANGO2, C22orf25 | transport and golgi organization 2 homolog (Drosophila) |
C00044491
|
| 9344 | TAOK2, MAP3K17, PSK, PSK1, PSK1-BETA, TAO1, TAO2 | TAO kinase 2 (EC:2.7.11.1) |
C00044491
|
| 6890 | TAP1, ABC17, ABCB2, APT1, D6S114E, PSF-1, PSF1, RING4, TAP1*0102N, TAP1N | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
C00044491
|
| 202018 | TAPT1, CMVFR | transmembrane anterior posterior transformation 1 |
C00044491
|
| 123283 | TARSL2 | threonyl-tRNA synthetase-like 2 (EC:6.1.1.3) |
C00044491
|
| 23232 | TBC1D12 | TBC1 domain family, member 12 |
C00044491
|
| 128637 | TBC1D20, C20orf140 | TBC1 domain family, member 20 |
C00044491
|
| 25771 | TBC1D22A, C22orf4, HSC79E021 | TBC1 domain family, member 22A |
C00044491
|
| 55633 | TBC1D22B, C6orf197, RP4-744I24.2, dJ744I24.2 | TBC1 domain family, member 22B |
C00044491
|
| 23102 | TBC1D2B | TBC1 domain family, member 2B |
C00044491
|
| 93594 | TBC1D31, Gm85, WDR67 | TBC1 domain family, member 31 |
C00044491
|
| 9779 | TBC1D5 | TBC1 domain family, member 5 |
C00044491
|
| 51256 | TBC1D7, PIG51, TBC7, dJ257A7.3 | TBC1 domain family, member 7 |
C00044491
|
| 11138 | TBC1D8, AD3, HBLP1, TBC1D8A, VRP | TBC1 domain family, member 8 (with GRAM domain) |
C00044491
|
| 54885 | TBC1D8B | TBC1 domain family, member 8B (with GRAM domain) |
C00044491
|
| 23158 | TBC1D9, MDR1 | TBC1 domain family, member 9 (with GRAM domain) |
C00044491
|
| 55171 | TBCCD1 | TBCC domain containing 1 |
C00044491
|
| 93627 | TBCK, TBCKL | TBC1 domain containing kinase |
C00044491
|
| 6907 | TBL1X, EBI, SMAP55, TBL1 | transducin (beta)-like 1X-linked |
C00044491
|
| 79718 | TBL1XR1, C21, DC42, IRA1, TBLR1 | transducin (beta)-like 1 X-linked receptor 1 |
C00044491
|
| 10607 | TBL3, SAZD, UTP13 | transducin (beta)-like 3 |
C00044491
|
| 9519 | TBPL1, MGC:8389, MGC:9620, STUD, TLF, TLP, TRF2 | TBP-like 1 |
C00044491
|
| 387332 | TBPL2, TBP2, TRF3 | TATA box binding protein like 2 |
C00044491
|
| 9238 | TBRG4, CPR2, FASTKD4 | transforming growth factor beta regulator 4 |
C00044491
|
| 30009 | TBX21, T-PET, T-bet, TBET, TBLYM | T-box 21 |
C00044491
|
| 285343 | TCAIM, C3orf23, TOAG-1, TOAG1 | T cell activation inhibitor, mitochondrial |
C00044491
|
| 6919 | TCEA2, TFIIS | transcription elongation factor A (SII), 2 |
C00044491
|
| 6920 | TCEA3, TFIIS, TFIIS.H | transcription elongation factor A (SII), 3 |
C00044491
|
| 90843 | TCEAL8 | transcription elongation factor A (SII)-like 8 |
C00044491
|
| 6921 | TCEB1, SIII, eloC | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
C00044491
|
| 6938 | TCF12, CRS3, HEB, HTF4, HsT17266, bHLHb20 | transcription factor 12 |
C00044491
|
| 6941 | TCF19, SC1, TCF-19 | transcription factor 19 |
C00044491
|
| 6929 | TCF3, E2A, E47, ITF1, TCF-3, VDIR, bHLHb21 | transcription factor 3 |
C00044491
|
| 6932 | TCF7, TCF-1 | transcription factor 7 (T-cell specific, HMG-box) |
C00044491
|
| 83439 | TCF7L1, TCF-3, TCF3 | transcription factor 7-like 1 (T-cell specific, HMG-box) |
C00044491
|
| 6934 | TCF7L2, TCF-4, TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) |
C00044491
|
| 6950 | TCP1, CCT-alpha, CCT1, CCTa, D6S230E, TCP-1-alpha | t-complex 1 |
C00044491
|
| 55346 | TCP11L1, dJ85M6.3 | t-complex 11, testis-specific-like 1 |
C00044491
|
| 255394 | TCP11L2 | t-complex 11, testis-specific-like 2 |
C00044491
|
| 6988 | TCTA | T-cell leukemia translocation altered |
C00044491
|
| 6996 | TDG, hTDG | thymine-DNA glycosylase (EC:3.2.2.29) |
C00044491
|
| 55775 | TDP1 | tyrosyl-DNA phosphodiesterase 1 (EC:3.1.4.-) |
C00044491
|
| 81550 | TDRD3 | tudor domain containing 3 |
C00044491
|
| 23424 | TDRD7, CATC4, CTRCT36, PCTAIRE2BP, RP11-508D10.1, TRAP | tudor domain containing 7 |
C00044491
|
| 11022 | TDRKH, TDRD2 | tudor and KH domain containing |
C00044491
|
| 7008 | TEF | thyrotrophic embryonic factor |
C00044491
|
| 79736 | TEFM, C17orf42 | transcription elongation factor, mitochondrial |
C00044491
|
| 23371 | TENC1, C1-TEN, C1TEN, TNS2 | tensin like C1 domain containing phosphatase (tensin 2) |
C00044491
|
| 7011 | TEP1, TLP1, TP1, TROVE1, VAULT2, p240 | telomerase-associated protein 1 |
C00044491
|
| 54386 | TERF2IP, DRIP5, RAP1 | telomeric repeat binding factor 2, interacting protein |
C00044491
|
| 26136 | TES, TESS, TESS-2 | testis derived transcript (3 LIM domains) |
C00044491
|
| 7016 | TESK1 | testis-specific kinase 1 (EC:2.7.12.1) |
C00044491
|
| 10420 | TESK2 | testis-specific kinase 2 (EC:2.7.12.1) |
C00044491
|
| 122046 | TEX26, C13orf26 | testis expressed 26 |
C00044491
|
| 113419 | TEX261, TEG-261 | testis expressed 261 |
C00044491
|
| 121793 | TEX29, C13orf16, bA474D23.1 | testis expressed 29 |
C00044491
|
| 93081 | TEX30, C13orf27 | testis expressed 30 |
C00044491
|
| 374618 | TEX9 | testis expressed 9 |
C00044491
|
| 7018 | TF, PRO1557, PRO2086, TFQTL1 | transferrin |
C00044491
|
| 7023 | TFAP4, AP-4, bHLHc41 | transcription factor AP-4 (activating enhancer binding protein 4) |
C00044491
|
| 51106 | TFB1M, CGI75, mtTFB, mtTFB1 | transcription factor B1, mitochondrial (EC:2.1.1.-) |
C00044491
|
| 64216 | TFB2M, Hkp1, mtTFB2 | transcription factor B2, mitochondrial |
C00044491
|
| 7027 | TFDP1, DP1, DRTF1, Dp-1 | transcription factor Dp-1 |
C00044491
|
| 10342 | TFG, HMSNP, SPG57, TF6, TRKT3 | TRK-fused gene |
C00044491
|
| 7035 | TFPI, EPI, LACI, TFI, TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
C00044491
|
| 7980 | TFPI2, PP5, REF1, TFPI-2 | tissue factor pathway inhibitor 2 |
C00044491
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00044491
|
| 7045 | TGFBI, BIGH3, CDB1, CDG2, CDGG1, CSD, CSD1, CSD2, CSD3, EBMD, LCD1 | transforming growth factor, beta-induced, 68kDa |
C00044491
|
| 7046 | TGFBR1, AAT5, ACVRLK4, ALK-5, ALK5, LDS1A, LDS2A, MSSE, SKR4, TGFR-1 | transforming growth factor, beta receptor 1 (EC:2.7.11.30) |
C00044491
|
| 7050 | TGIF1, HPE4, TGIF | TGFB-induced factor homeobox 1 |
C00044491
|
| 60436 | TGIF2 | TGFB-induced factor homeobox 2 |
C00044491
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00044491
|
| 96764 | TGS1, NCOA6IP, PIMT, PIPMT | trimethylguanosine synthase 1 (EC:2.1.1.-) |
C00044491
|
| 63892 | THADA, GITA | thyroid adenoma associated |
C00044491
|
| 7056 | THBD, AHUS6, BDCA3, CD141, THPH12, THRM, TM | thrombomodulin |
C00044491
|
| 7057 | THBS1, THBS, THBS-1, TSP, TSP-1, TSP1 | thrombospondin 1 |
C00044491
|
| 79896 | THNSL1, TSH1 | threonine synthase-like 1 (S. cerevisiae) |
C00044491
|
| 80145 | THOC7, NIF3L1BP1, fSAP24, hTREX30 | THO complex 7 homolog (Drosophila) |
C00044491
|
| 7066 | THPO, MGDF, MKCSF, ML, MPLLG, THCYT1, TPO | thrombopoietin |
C00044491
|
| 7069 | THRSP, LPGP1, Lpgp, S14, SPOT14, THRP | thyroid hormone responsive |
C00044491
|
| 80731 | THSD7B | thrombospondin, type I, domain containing 7B |
C00044491
|
| 29087 | THYN1, MDS012, MY105, THY28, THY28KD | thymocyte nuclear protein 1 |
C00044491
|
| 7072 | TIA1, TIA-1, WDM | TIA1 cytotoxic granule-associated RNA binding protein |
C00044491
|
| 90381 | TICRR, C15orf42, SLD3, Treslin | TOPBP1-interacting checkpoint and replication regulator |
C00044491
|
| 7075 | TIE1, JTK14, TIE | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 (EC:2.7.10.1) |
C00044491
|
| 8914 | TIMELESS, TIM, TIM1, hTIM | timeless circadian clock |
C00044491
|
| 10440 | TIMM17A, TIM17, TIM17A | translocase of inner mitochondrial membrane 17 homolog A (yeast) |
C00044491
|
| 29090 | TIMM21, C18orf55, TIM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) |
C00044491
|
| 7076 | TIMP1, CLGI, EPA, EPO, HCI, TIMP | TIMP metallopeptidase inhibitor 1 |
C00044491
|
| 7077 | TIMP2, CSC-21K, DDC8 | TIMP metallopeptidase inhibitor 2 |
C00044491
|
| 7078 | TIMP3, HSMRK222, K222, K222TA2, SFD | TIMP metallopeptidase inhibitor 3 |
C00044491
|
| 7079 | TIMP4 | TIMP metallopeptidase inhibitor 4 |
C00044491
|
| 64129 | TINAGL1, ARG1, LCN7, LIECG3, TINAGRP | tubulointerstitial nephritis antigen-like 1 |
C00044491
|
| 25976 | TIPARP, ARTD14, PARP7, pART14 | TCDD-inducible poly(ADP-ribose) polymerase (EC:2.4.2.30) |
C00044491
|
| 9414 | TJP2, C9DUPq21.11, DFNA51, DUP9q21.11, X104, ZO2 | tight junction protein 2 |
C00044491
|
| 27134 | TJP3, ZO-3, ZO3 | tight junction protein 3 |
C00044491
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00044491
|
| 7084 | TK2, MTDPS2, MTTK | thymidine kinase 2, mitochondrial (EC:2.7.1.21) |
C00044491
|
| 7086 | TKT, TK, TKT1 | transketolase (EC:2.2.1.1) |
C00044491
|
| 7088 | TLE1, ESG, ESG1, GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
C00044491
|
| 7090 | TLE3, ESG, ESG3, GRG3, HsT18976 | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
C00044491
|
| 9874 | TLK1, PKU-beta | tousled-like kinase 1 (EC:2.7.11.1) |
C00044491
|
| 7096 | TLR1, CD281, TIL, TIL._LPRS5, rsc786 | toll-like receptor 1 |
C00044491
|
| 7098 | TLR3, CD283, IIAE2 | toll-like receptor 3 |
C00044491
|
| 83877 | TM2D2, BLP1 | TM2 domain containing 2 |
C00044491
|
| 4071 | TM4SF1, H-L6, L6, M3S1, TAAL6 | transmembrane 4 L six family member 1 |
C00044491
|
| 116441 | TM4SF18, L6D | transmembrane 4 L six family member 18 |
C00044491
|
| 7104 | TM4SF4, ILTMP, il-TMP | transmembrane 4 L six family member 4 |
C00044491
|
| 51768 | TM7SF3 | transmembrane 7 superfamily member 3 |
C00044491
|
| 10548 | TM9SF1, HMP70, MP70 | transmembrane 9 superfamily member 1 |
C00044491
|
| 9375 | TM9SF2, P76 | transmembrane 9 superfamily member 2 |
C00044491
|
| 56889 | TM9SF3, EP70-P-iso, SMBP | transmembrane 9 superfamily member 3 |
C00044491
|
| 79905 | TMC7 | transmembrane channel-like 7 |
C00044491
|
| 23023 | TMCC1 | transmembrane and coiled-coil domain family 1 |
C00044491
|
| 55002 | TMCO3, C13orf11 | transmembrane and coiled-coil domains 3 |
C00044491
|
| 55374 | TMCO6 | transmembrane and coiled-coil domains 6 |
C00044491
|
| 222068 | TMED4, ERS25, HNLF | transmembrane emp24 protein transport domain containing 4 |
C00044491
|
| 54868 | TMEM104 | transmembrane protein 104 |
C00044491
|
| 79022 | TMEM106C | transmembrane protein 106C |
C00044491
|
| 84314 | TMEM107, GRVS638, PRO1268 | transmembrane protein 107 |
C00044491
|
| 11070 | TMEM115, PL6 | transmembrane protein 115 |
C00044491
|
| 89894 | TMEM116 | transmembrane protein 116 |
C00044491
|
| 80757 | TMEM121, hole | transmembrane protein 121 |
C00044491
|
| 128218 | TMEM125 | transmembrane protein 125 |
C00044491
|
| 83935 | TMEM133 | transmembrane protein 133 |
C00044491
|
| 65084 | TMEM135, PMP52 | transmembrane protein 135 |
C00044491
|
| 219902 | TMEM136 | transmembrane protein 136 |
C00044491
|
| 51524 | TMEM138 | transmembrane protein 138 |
C00044491
|
| 129303 | TMEM150A, TM6P1, TMEM150 | transmembrane protein 150A |
C00044491
|
| 153396 | TMEM161B, FLB3342, PRO1313 | transmembrane protein 161B |
C00044491
|
| 124491 | TMEM170A, TMEM170 | transmembrane protein 170A |
C00044491
|
| 84286 | TMEM175 | transmembrane protein 175 |
C00044491
|
| 28959 | TMEM176B, LR8 | transmembrane protein 176B |
C00044491
|
| 25829 | TMEM184B, C22orf5, FM08, HS5O6A | transmembrane protein 184B |
C00044491
|
| 79134 | TMEM185B, FAM11B | transmembrane protein 185B |
C00044491
|
| 55266 | TMEM19 | transmembrane protein 19 |
C00044491
|
| 23306 | TMEM194A, TMEM194 | transmembrane protein 194A |
C00044491
|
| 100131211 | TMEM194B | transmembrane protein 194B |
C00044491
|
| 199953 | TMEM201, NET5 | transmembrane protein 201 |
C00044491
|
| 94107 | TMEM203, HBEBP1 | transmembrane protein 203 |
C00044491
|
| 84928 | TMEM209, NET31 | transmembrane protein 209 |
C00044491
|
| 79064 | TMEM223 | transmembrane protein 223 |
C00044491
|
| 29058 | TMEM230, C20orf30, HSPC274, dJ1116H23.2.1 | transmembrane protein 230 |
C00044491
|
| 79161 | TMEM243, C7orf23, MM-TRAG | transmembrane protein 243, mitochondrial |
C00044491
|
| 23731 | TMEM245, C9orf5, CG-2, CG2 | transmembrane protein 245 |
C00044491
|
| 26175 | TMEM251, C14orf109 | transmembrane protein 251 |
C00044491
|
| 57393 | TMEM27, NX-17, NX17 | transmembrane protein 27 |
C00044491
|
| 55754 | TMEM30A, C6orf67, CDC50A | transmembrane protein 30A |
C00044491
|
| 55151 | TMEM38B, C9orf87, D4Ertd89e, OI14, TRIC-B, TRICB, bA219P18.1 | transmembrane protein 38B |
C00044491
|
| 90407 | TMEM41A, 2900010K02Rik | transmembrane protein 41A |
C00044491
|
| 55076 | TMEM45A, DERP7 | transmembrane protein 45A |
C00044491
|
| 83604 | TMEM47, BCMP1, TM4SF10 | transmembrane protein 47 |
C00044491
|
| 23585 | TMEM50A, IFNRC, SMP1 | transmembrane protein 50A |
C00044491
|
| 757 | TMEM50B, C21orf4, HCVP7TP3 | transmembrane protein 50B |
C00044491
|
| 55092 | TMEM51, C1orf72 | transmembrane protein 51 |
C00044491
|
| 113452 | TMEM54, BCLP, CAC-1, CAC1 | transmembrane protein 54 |
C00044491
|
| 90809 | TMEM55B, C14orf9 | transmembrane protein 55B (EC:3.1.3.78) |
C00044491
|
| 9528 | TMEM59, C1orf8, HSPC001 | transmembrane protein 59 |
C00044491
|
| 25789 | TMEM59L, BSMAP, C19orf4 | transmembrane protein 59-like |
C00044491
|
| 85025 | TMEM60, C7orf35 | transmembrane protein 60 |
C00044491
|
| 80021 | TMEM62 | transmembrane protein 62 |
C00044491
|
| 55362 | TMEM63B, C6orf110, RP3-421H19.2 | transmembrane protein 63B |
C00044491
|
| 157378 | TMEM65 | transmembrane protein 65 |
C00044491
|
| 51669 | TMEM66, FOAP-7, SARAF, XTP3 | transmembrane protein 66 |
C00044491
|
| 137695 | TMEM68 | transmembrane protein 68 |
C00044491
|
| 51249 | TMEM69, C1orf154, RP11-767N6.4 | transmembrane protein 69 |
C00044491
|
| 54968 | TMEM70, MC5DN2 | transmembrane protein 70 |
C00044491
|
| 84910 | TMEM87B | transmembrane protein 87B |
C00044491
|
| 440955 | TMEM89 | transmembrane protein 89 |
C00044491
|
| 27346 | TMEM97, MAC30 | transmembrane protein 97 |
C00044491
|
| 56674 | TMEM9B, C11orf15 | TMEM9 domain family, member B |
C00044491
|
| 7110 | TMF1, ARA160, TMF | TATA element modulatory factor 1 |
C00044491
|
| 126259 | TMIGD2, IGPR-1, IGPR1 | transmembrane and immunoglobulin domain containing 2 |
C00044491
|
| 29766 | TMOD3, UTMOD | tropomodulin 3 (ubiquitous) |
C00044491
|
| 7112 | TMPO, CMD1T, LAP2, LEMD4, PRO0868, TP | thymopoietin |
C00044491
|
| 84000 | TMPRSS13, MSP, MSPL, MSPS, TMPRSS11 | transmembrane protease, serine 13 |
C00044491
|
| 7113 | TMPRSS2, PP9284, PRSS10 | transmembrane protease, serine 2 (EC:3.4.21.-) |
C00044491
|
| 64699 | TMPRSS3, DFNB10, DFNB8, ECHOS1, TADG12 | transmembrane protease, serine 3 (EC:3.4.21.-) |
C00044491
|
| 160418 | TMTC3, SMILE | transmembrane and tetratricopeptide repeat containing 3 |
C00044491
|
| 84899 | TMTC4 | transmembrane and tetratricopeptide repeat containing 4 |
C00044491
|
| 81542 | TMX1, PDIA11, TMX, TXNDC, TXNDC1 | thioredoxin-related transmembrane protein 1 |
C00044491
|
| 7126 | TNFAIP1, B12, B61, BTBD34, EDP1, hBACURD2 | tumor necrosis factor, alpha-induced protein 1 (endothelial) |
C00044491
|
| 126282 | TNFAIP8L1, TIPE1 | tumor necrosis factor, alpha-induced protein 8-like 1 |
C00044491
|
| 8795 | TNFRSF10B, CD262, DR5, KILLER, KILLER/DR5, TRAIL-R2, TRAILR2, TRICK2, TRICK2A, TRICK2B, TRICKB, ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b |
C00044491
|
| 8793 | TNFRSF10D, CD264, DCR2, TRAIL-R4, TRAILR4, TRUNDD | tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
C00044491
|
| 4982 | TNFRSF11B, OCIF, OPG, TR1 | tumor necrosis factor receptor superfamily, member 11b |
C00044491
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00044491
|
| 7132 | TNFRSF1A, CD120a, FPF, MS5, TBP1, TNF-R, TNF-R-I, TNF-R55, TNFAR, TNFR1, TNFR1-d2, TNFR55, TNFR60, p55, p55-R, p60 | tumor necrosis factor receptor superfamily, member 1A |
C00044491
|
| 7293 | TNFRSF4, ACT35, CD134, OX40, TXGP1L | tumor necrosis factor receptor superfamily, member 4 |
C00044491
|
| 943 | TNFRSF8, CD30, D1S166E, Ki-1 | tumor necrosis factor receptor superfamily, member 8 |
C00044491
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00044491
|
| 8600 | TNFSF11, CD254, ODF, OPGL, OPTB2, RANKL, TRANCE, hRANKL2, sOdf | tumor necrosis factor (ligand) superfamily, member 11 |
C00044491
|
| 8741 | TNFSF13, APRIL, CD256, TALL-2, TALL2, TRDL-1, ZTNF2 | tumor necrosis factor (ligand) superfamily, member 13 |
C00044491
|
| 7140 | TNNT3, TNTF | troponin T type 3 (skeletal, fast) |
C00044491
|
| 27320 |
C00044491
|
||
| 27327 | TNRC6A, CAGH26, GW1, GW182, TNRC6 | trinucleotide repeat containing 6A |
C00044491
|
| 23112 | TNRC6B | trinucleotide repeat containing 6B |
C00044491
|
| 57690 | TNRC6C | trinucleotide repeat containing 6C |
C00044491
|
| 7145 | TNS1, MST091, MST122, MST127, MSTP122, MSTP127, MXRA6, TNS | tensin 1 |
C00044491
|
| 64759 | TNS3, TEM6, TENS1 | tensin 3 |
C00044491
|
| 84951 | TNS4, CTEN | tensin 4 |
C00044491
|
| 146691 | TOM1L2 | target of myb1-like 2 (chicken) |
C00044491
|
| 56993 | TOMM22, 1C9-2, MST065, MSTP065, TOM22 | translocase of outer mitochondrial membrane 22 homolog (yeast) |
C00044491
|
| 84134 | TOMM40L, RP11-297K8.10, TOMM40B | translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
C00044491
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00044491
|
| 7155 | TOP2B, TOPIIB, top2beta | topoisomerase (DNA) II beta 180kDa (EC:5.99.1.3) |
C00044491
|
| 11073 | TOPBP1, TOP2BP1 | topoisomerase (DNA) II binding protein 1 |
C00044491
|
| 27348 | TOR1B, DQ1 | torsin family 1, member B (torsin B) |
C00044491
|
| 64222 | TOR3A, ADIR, ADIR2 | torsin family 3, member A |
C00044491
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00044491
|
| 7159 | TP53BP2, 53BP2, ASPP2, BBP, P53BP2, PPP1R13A | tumor protein p53 binding protein, 2 |
C00044491
|
| 9540 | TP53I3, PIG3 | tumor protein p53 inducible protein 3 |
C00044491
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00044491
|
| 58476 | TP53INP2, C20orf110, DOR, PINH, dJ1181N3.1 | tumor protein p53 inducible nuclear protein 2 |
C00044491
|
| 11257 | TP53TG1, LINC00096, NCRNA00096, P53TG1, P53TG1-D, TP53AP1 | TP53 target 1 (non-protein coding) |
C00044491
|
| 53373 | TPCN1, TPC1 | two pore segment channel 1 |
C00044491
|
| 7164 | TPD52L1, D53, hD53 | tumor protein D52-like 1 |
C00044491
|
| 7168 | TPM1, C15orf13, CMD1Y, CMH3, HTM-alpha, LVNC9, TMSA | tropomyosin 1 (alpha) |
C00044491
|
| 7171 | TPM4 | tropomyosin 4 |
C00044491
|
| 7174 | TPP2, TPP-2 | tripeptidyl peptidase II (EC:3.4.14.10) |
C00044491
|
| 25823 | TPSG1, PRSS31, TMT, trpA | tryptase gamma 1 (EC:3.4.21.59) |
C00044491
|
| 29896 | TRA2A, AWMS1, HSU53209 | transformer 2 alpha homolog (Drosophila) |
C00044491
|
| 7186 | TRAF2, MGC:45012, TRAP, TRAP3 | TNF receptor-associated factor 2 |
C00044491
|
| 7187 | TRAF3, CAP-1, CAP1, CD40bp, CRAF1, IIAE5, LAP1 | TNF receptor-associated factor 3 |
C00044491
|
| 10758 | TRAF3IP2, ACT1, C6orf2, C6orf4, C6orf5, C6orf6, CIKS, PSORS13 | TRAF3 interacting protein 2 |
C00044491
|
| 9618 | TRAF4, CART1, MLN62, RNF83 | TNF receptor-associated factor 4 |
C00044491
|
| 10906 | TRAFD1, FLN29 | TRAF-type zinc finger domain containing 1 |
C00044491
|
| 10293 | TRAIP, RNF206, TRIP | TRAF interacting protein |
C00044491
|
| 22906 | TRAK1, MILT1, OIP106 | trafficking protein, kinesin binding 1 |
C00044491
|
| 66008 | TRAK2, ALS2CR3, CALS-C, GRIF-1, GRIF1, MILT2, OIP98 | trafficking protein, kinesin binding 2 |
C00044491
|
| 9697 | TRAM2 | translocation associated membrane protein 2 |
C00044491
|
| 10131 | TRAP1, HSP_75, HSP75, HSP90L, TRAP-1 | TNF receptor-associated protein 1 |
C00044491
|
| 58485 | TRAPPC1, BET5, MUM2 | trafficking protein particle complex 1 |
C00044491
|
| 7109 | TRAPPC10, EHOC-1, EHOC1, GT334, TMEM1, TRS130, TRS30 | trafficking protein particle complex 10 |
C00044491
|
| 51112 | TRAPPC12, TTC-15, TTC15 | trafficking protein particle complex 12 |
C00044491
|
| 83696 | TRAPPC9, IBP, IKBKBBP, MRT13, NIBP, TRS120 | trafficking protein particle complex 9 |
C00044491
|
| 51499 | TRIAP1, MDM35, P53CSV, WF-1 | TP53 regulated inhibitor of apoptosis 1 |
C00044491
|
| 28951 | TRIB2, C5FW, GS3955, TRB2 | tribbles pseudokinase 2 |
C00044491
|
| 57761 | TRIB3, C20orf97, NIPK, SINK, SKIP3, TRB3 | tribbles pseudokinase 3 |
C00044491
|
| 9830 | TRIM14 | tripartite motif containing 14 |
C00044491
|
| 6737 | TRIM21, RNF81, RO52, SSA, SSA1 | tripartite motif containing 21 |
C00044491
|
| 10346 | TRIM22, GPSTAF50, RNF94, STAF50 | tripartite motif containing 22 |
C00044491
|
| 8805 | TRIM24, PTC6, RNF82, TF1A, TIF1, TIF1A, TIF1ALPHA, hTIF1 | tripartite motif containing 24 |
C00044491
|
| 7706 | TRIM25, EFP, RNF147, Z147, ZNF147 | tripartite motif containing 25 (EC:6.3.2.19 6.3.2.n3) |
C00044491
|
| 7726 | TRIM26, AFP, RNF95, ZNF173 | tripartite motif containing 26 |
C00044491
|
| 5987 | TRIM27, RFP, RNF76 | tripartite motif containing 27 |
C00044491
|
| 22954 | TRIM32, BBS11, HT2A, LGMD2H, TATIP | tripartite motif containing 32 |
C00044491
|
| 51592 | TRIM33, ECTO, PTC7, RFG7, TF1G, TIF1G, TIF1GAMMA, TIFGAMMA | tripartite motif containing 33 |
C00044491
|
| 23087 | TRIM35, HLS5, MAIR | tripartite motif containing 35 |
C00044491
|
| 10475 | TRIM38, RNF15, RORET | tripartite motif containing 38 |
C00044491
|
| 90933 | TRIM41, RINCK | tripartite motif containing 41 |
C00044491
|
| 54765 | TRIM44, DIPB, HSA249128, MC7 | tripartite motif containing 44 |
C00044491
|
| 91107 | TRIM47, GOA, RNF100 | tripartite motif containing 47 |
C00044491
|
| 85363 | TRIM5, RNF88, TRIM5alpha | tripartite motif containing 5 |
C00044491
|
| 140691 | TRIM69, HSD34, RNF36, Trif | tripartite motif containing 69 |
C00044491
|
| 81603 | TRIM8, GERP, RNF27 | tripartite motif containing 8 |
C00044491
|
| 11078 | TRIOBP, DFNB28, TARA, dJ37E16.4 | TRIO and F-actin binding protein |
C00044491
|
| 9321 | TRIP11, ACG1A, CEV14, GMAP-210, TRIP-11, TRIP230 | thyroid hormone receptor interactor 11 |
C00044491
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00044491
|
| 9325 | TRIP4, ASC-1, ASC1, HsT17391 | thyroid hormone receptor interactor 4 |
C00044491
|
| 7205 | TRIP6, OIP-1, OIP1, TRIP-6, TRIP6i2, ZRP-1 | thyroid hormone receptor interactor 6 |
C00044491
|
| 158234 | TRMT10B, RG9MTD3, RP11-3J10.9, bA3J10.9 | tRNA methyltransferase 10 homolog B (S. cerevisiae) |
C00044491
|
| 54931 | TRMT10C, HNYA, MRPP1, RG9MTD1 | tRNA methyltransferase 10 homolog C (S. cerevisiae) |
C00044491
|
| 54482 | TRMT13, CCDC76 | tRNA methyltransferase 13 homolog (S. cerevisiae) (EC:2.1.1.225) |
C00044491
|
| 81627 | TRMT1L, C1orf25, MST070, TRM1L, bG120K12.3 | tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
C00044491
|
| 55006 | TRMT61B | tRNA methyltransferase 61 homolog B (S. cerevisiae) (EC:2.1.1.220) |
C00044491
|
| 54952 | TRNAU1AP, PRO1902, RP4-669K10.4, SECP43, TRSPAP1 | tRNA selenocysteine 1 associated protein 1 |
C00044491
|
| 388610 | TRNP1, C1orf225, TNRP | TMF1-regulated nuclear protein 1 |
C00044491
|
| 10024 | TROAP, TASTIN | trophinin associated protein |
C00044491
|
| 7220 | TRPC1, HTRP-1, TRP1 | transient receptor potential cation channel, subfamily C, member 1 |
C00044491
|
| 29850 | TRPM5, LTRPC5, MTR1 | transient receptor potential cation channel, subfamily M, member 5 |
C00044491
|
| 79054 | TRPM8, LTRPC6, TRPP8 | transient receptor potential cation channel, subfamily M, member 8 |
C00044491
|
| 59341 | TRPV4, CMT2C, HMSN2C, OTRPC4, SMAL, SPSMA, SSQTL1, TRP12, VRL2, VROAC | transient receptor potential cation channel, subfamily V, member 4 |
C00044491
|
| 8295 | TRRAP, PAF350/400, PAF400, STAF40, TR-AP, Tra1 | transformation/transcription domain-associated protein |
C00044491
|
| 142940 | TRUB1, PUS4 | TruB pseudouridine (psi) synthase family member 1 |
C00044491
|
| 7249 | TSC2, LAM, TSC4 | tuberous sclerosis 2 |
C00044491
|
| 9819 | TSC22D2, TILZ4a, TILZ4b, TILZ4c | TSC22 domain family, member 2 |
C00044491
|
| 1831 | TSC22D3, DIP, DSIPI, GILZ, TSC-22R, hDIP | TSC22 domain family, member 3 |
C00044491
|
| 80705 | TSGA10, CEP4L, CT79 | testis specific, 10 |
C00044491
|
| 85480 | TSLP | thymic stromal lymphopoietin |
C00044491
|
| 7247 | TSN, BCLF-1, C3PO, RCHF1, REHF-1, TBRBP, TRSLN | translin |
C00044491
|
| 203062 | TSNARE1 | t-SNARE domain containing 1 |
C00044491
|
| 23554 | TSPAN12, EVR5, NET-2, NET2, TM4SF12 | tetraspanin 12 |
C00044491
|
| 27075 | TSPAN13, NET-6, NET6, TM4SF13 | tetraspanin 13 |
C00044491
|
| 10099 | TSPAN3, TM4-A, TM4SF8, TSPAN-3 | tetraspanin 3 |
C00044491
|
| 6302 | TSPAN31, SAS | tetraspanin 31 |
C00044491
|
| 340348 | TSPAN33, PEN | tetraspanin 33 |
C00044491
|
| 7106 | TSPAN4, NAG-2, NAG2, TETRASPAN, TM4SF7, TSPAN-4 | tetraspanin 4 |
C00044491
|
| 7103 | TSPAN8, CO-029, TM4SF3 | tetraspanin 8 |
C00044491
|
| 7259 | TSPYL1, TSPYL | TSPY-like 1 |
C00044491
|
| 241732 |
C00044491
|
||
| 158427 | TSTD2, C9orf97 | thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
C00044491
|
| 151613 | TTC14, DRDL5813, PRO19630 | tetratricopeptide repeat domain 14 |
C00044491
|
| 55761 | TTC17 | tetratricopeptide repeat domain 17 |
C00044491
|
| 55622 | TTC27 | tetratricopeptide repeat domain 27 |
C00044491
|
| 23331 | TTC28, TPRBK | tetratricopeptide repeat domain 28 |
C00044491
|
| 7267 | TTC3, DCRR1, RNF105, TPRDIII | tetratricopeptide repeat domain 3 |
C00044491
|
| 64427 | TTC31 | tetratricopeptide repeat domain 31 |
C00044491
|
| 55020 | TTC38 | tetratricopeptide repeat domain 38 |
C00044491
|
| 125488 | TTC39C, C18orf17, HsT2697 | tetratricopeptide repeat domain 39C |
C00044491
|
| 7268 | TTC4 | tetratricopeptide repeat domain 4 |
C00044491
|
| 57217 | TTC7A, MINAT, TTC7 | tetratricopeptide repeat domain 7A |
C00044491
|
| 8458 | TTF2, HuF2 | transcription termination factor, RNA polymerase II |
C00044491
|
| 9675 | TTI1, KIAA0406, smg-10 | TELO2 interacting protein 1 |
C00044491
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00044491
|
| 150465 | TTL | tubulin tyrosine ligase (EC:6.3.2.25) |
C00044491
|
| 25809 | TTLL1, C22orf7, HS323M22B | tubulin tyrosine ligase-like family, member 1 |
C00044491
|
| 254173 | TTLL10, TTLL5 | tubulin tyrosine ligase-like family, member 10 (EC:6.-.-.-) |
C00044491
|
| 23170 | TTLL12, dJ526I14.2 | tubulin tyrosine ligase-like family, member 12 |
C00044491
|
| 26140 | TTLL3, HOTTL | tubulin tyrosine ligase-like family, member 3 |
C00044491
|
| 79739 | TTLL7, RP5-836J3.2 | tubulin tyrosine ligase-like family, member 7 |
C00044491
|
| 7274 | TTPA, ATTP, AVED, TTP1, alphaTTP | tocopherol (alpha) transfer protein |
C00044491
|
| 79183 | TTPAL, C20orf121 | tocopherol (alpha) transfer protein-like |
C00044491
|
| 84790 | TUBA1C, TUBA6, bcm948 | tubulin, alpha 1c |
C00044491
|
| 7278 | TUBA3C, TUBA2, bA408E5.3 | tubulin, alpha 3c |
C00044491
|
| 7277 | TUBA4A, H2-ALPHA, TUBA1 | tubulin, alpha 4a |
C00044491
|
| 100691897 |
C00044491
|
||
| 84617 | TUBB6, HsT1601, TUBB-5 | tubulin, beta 6 class V |
C00044491
|
| 10426 | TUBGCP3, 104p, GCP3, Grip104, SPBC98, Spc98, Spc98p | tubulin, gamma complex associated protein 3 |
C00044491
|
| 114791 | TUBGCP5, GCP5 | tubulin, gamma complex associated protein 5 |
C00044491
|
| 7286 | TUFT1 | tuftelin 1 |
C00044491
|
| 7289 | TULP3, TUBL3 | tubby like protein 3 |
C00044491
|
| 57045 | TWSG1, TSG | twisted gastrulation BMP signaling modulator 1 |
C00044491
|
| 7295 | TXN, TRDX, TRX, TRX1 | thioredoxin |
C00044491
|
| 57544 | TXNDC16, ERp90, KIAA1344 | thioredoxin domain containing 16 |
C00044491
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00044491
|
| 7297 | TYK2, JTK1 | tyrosine kinase 2 (EC:2.7.10.2) |
C00044491
|
| 1890 | TYMP, ECGF, ECGF1, MEDPS1, MNGIE, MTDPS1, PDECGF, TP, hPD-ECGF | thymidine phosphorylase (EC:2.4.2.4) |
C00044491
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00044491
|
| 7307 | U2AF1, FP793, RN, RNU2AF1, U2AF35, U2AFBP | U2 small nuclear RNA auxiliary factor 1 |
C00044491
|
| 23350 | U2SURP, SR140, fSAPa | U2 snRNP-associated SURP domain containing |
C00044491
|
| 55075 | UACA, NUCLING | uveal autoantigen with coiled-coil domains and ankyrin repeats |
C00044491
|
| 55236 | UBA6, E1-L2, MOP-4, UBE1L2 | ubiquitin-like modifier activating enzyme 6 |
C00044491
|
| 283991 | UBALD2, FAM100B | UBA-like domain containing 2 |
C00044491
|
| 55833 | UBAP2, UBAP-2 | ubiquitin associated protein 2 |
C00044491
|
| 7319 | UBE2A, HHR6A, MRXS30, MRXSN, RAD6A, UBC2 | ubiquitin-conjugating enzyme E2A (EC:6.3.2.19) |
C00044491
|
| 7320 | UBE2B, E2-17kDa, HHR6B, HR6B, RAD6B, UBC2 | ubiquitin-conjugating enzyme E2B (EC:6.3.2.19) |
C00044491
|
| 11065 | UBE2C, UBCH10, dJ447F3.2 | ubiquitin-conjugating enzyme E2C (EC:6.3.2.19) |
C00044491
|
| 7323 | UBE2D3, E2(17)KB3, UBC4/5, UBCH5C | ubiquitin-conjugating enzyme E2D 3 (EC:6.3.2.19) |
C00044491
|
| 7324 | UBE2E1, UBCH6 | ubiquitin-conjugating enzyme E2E 1 (EC:6.3.2.19) |
C00044491
|
| 7326 | UBE2G1, E217K, UBC7, UBE2G | ubiquitin-conjugating enzyme E2G 1 (EC:6.3.2.19) |
C00044491
|
| 7328 | UBE2H, E2-20K, GID3, UBC8, UBCH, UBCH2 | ubiquitin-conjugating enzyme E2H (EC:6.3.2.19) |
C00044491
|
| 3093 | UBE2K, E2-25K, HIP2, HYPG, LIG, UBC1 | ubiquitin-conjugating enzyme E2K (EC:6.3.2.19) |
C00044491
|
| 9246 | UBE2L6, RIG-B, UBCH8 | ubiquitin-conjugating enzyme E2L 6 (EC:6.3.2.19) |
C00044491
|
| 389898 | UBE2NL, Li174 | ubiquitin-conjugating enzyme E2N-like (EC:6.3.2.-) |
C00044491
|
| 27338 | UBE2S, E2-EPF, E2EPF, EPF5 | ubiquitin-conjugating enzyme E2S (EC:6.3.2.19) |
C00044491
|
| 29089 | UBE2T, PIG50 | ubiquitin-conjugating enzyme E2T (putative) (EC:6.3.2.19) |
C00044491
|
| 65264 | UBE2Z, USE1 | ubiquitin-conjugating enzyme E2Z (EC:6.3.2.19) |
C00044491
|
| 89910 | UBE3B, BPIDS | ubiquitin protein ligase E3B (EC:6.3.2.-) |
C00044491
|
| 9690 | UBE3C, HECTH2 | ubiquitin protein ligase E3C |
C00044491
|
| 10277 | UBE4B, E4, HDNB1, UBOX3, UFD2, UFD2A | ubiquitination factor E4B |
C00044491
|
| 29979 | UBQLN1, DA41, DSK2, PLIC-1, UBQN, XDRP1 | ubiquilin 1 |
C00044491
|
| 197131 | UBR1, JBS | ubiquitin protein ligase E3 component n-recognin 1 |
C00044491
|
| 23304 | UBR2, C6orf133, RP3-392M17.3, bA49A4.1, dJ242G1.1, dJ392M17.3 | ubiquitin protein ligase E3 component n-recognin 2 |
C00044491
|
| 51366 | UBR5, DD5, EDD, EDD1, HYD | ubiquitin protein ligase E3 component n-recognin 5 |
C00044491
|
| 55148 | UBR7, C14orf130 | ubiquitin protein ligase E3 component n-recognin 7 (putative) |
C00044491
|
| 92181 | UBTD2, DCUBP | ubiquitin domain containing 2 |
C00044491
|
| 652995 | UCA1, CUDR, LINC00178, NCRNA00178, UCAT1 | urothelial cancer associated 1 (non-protein coding) |
C00044491
|
| 7345 | UCHL1, NDGOA, PARK5, PGP_9.5, PGP9.5, PGP95, Uch-L1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (EC:3.4.19.12) |
C00044491
|
| 7347 | UCHL3, UCH-L3 | ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) (EC:3.4.19.12) |
C00044491
|
| 90226 | UCN2, SRP, UCN-II, UCNI, UR, URP | urocortin 2 |
C00044491
|
| 55293 | UEVLD, ATTP, UEV3 | UEV and lactate/malate dehyrogenase domains |
C00044491
|
| 7353 | UFD1L, UFD1 | ubiquitin fusion degradation 1 like (yeast) |
C00044491
|
| 7357 | UGCG, GCS, GLCT1 | UDP-glucose ceramide glucosyltransferase (EC:2.4.1.80) |
C00044491
|
| 7358 | UGDH, GDH, UDP-GlcDH, UDPGDH, UGD | UDP-glucose 6-dehydrogenase (EC:1.1.1.22) |
C00044491
|
| 55757 | UGGT2, HUGT2, UGCGL2, UGT2 | UDP-glucose glycoprotein glucosyltransferase 2 |
C00044491
|
| 7360 | UGP2, UDPG, UDPGP, UDPGP2, UGP1, UGPP1, UGPP2, pHC379 | UDP-glucose pyrophosphorylase 2 (EC:2.7.7.9) |
C00044491
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00044491
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00044491
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00044491
|
| 54578 | UGT1A6, GNT1, HLUGP, HLUGP1, UDPGT, UDPGT_1-6, UGT1, UGT1A6S, UGT1F | UDP glucuronosyltransferase 1 family, polypeptide A6 (EC:2.4.1.17) |
C00044491
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00044491
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00044491
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00044491
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00044491
|
| 127933 | UHMK1, KIS, KIST, P-CIP2 | U2AF homology motif (UHM) kinase 1 (EC:2.7.11.1) |
C00044491
|
| 54887 | UHRF1BP1, C6orf107, ICBP90, dJ349A12.1 | UHRF1 binding protein 1 |
C00044491
|
| 80328 | ULBP2, N2DL2, RAET1H | UL16 binding protein 2 |
C00044491
|
| 25989 | ULK3 | unc-51 like kinase 3 (EC:2.7.11.1) |
C00044491
|
| 23025 | UNC13A, Munc13-1 | unc-13 homolog A (C. elegans) |
C00044491
|
| 55898 | UNC45A, GC-UNC45, GCUNC-45, GCUNC45, IRO039700, SMAP-1, SMAP1 | unc-45 homolog A (C. elegans) |
C00044491
|
| 222643 | UNC5CL, MUXA, ZUD | unc-5 homolog C (C. elegans)-like |
C00044491
|
| 7374 | UNG, DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 | uracil-DNA glycosylase (EC:3.2.2.27) |
C00044491
|
| 85451 | UNK, ZC3H5, ZC3HDC5 | unkempt family zinc finger |
C00044491
|
| 64718 | UNKL, C16orf28, ZC3H5L, ZC3HDC5L | unkempt family zinc finger-like |
C00044491
|
| 55245 | UQCC1, BFZB, C20orf44, CBP3, UQCC | ubiquinol-cytochrome c reductase complex assembly factor 1 |
C00044491
|
| 7385 | UQCRC2, MC3DN5, QCR2, UQCR2 | ubiquinol-cytochrome c reductase core protein II (EC:1.10.2.2) |
C00044491
|
| 55665 | URGCP, URG4 | upregulator of cell proliferation |
C00044491
|
| 131669 | UROC1, HMFN0320 | urocanate hydratase 1 (EC:4.2.1.49) |
C00044491
|
| 7389 | UROD, PCT, UPD | uroporphyrinogen decarboxylase (EC:4.1.1.37) |
C00044491
|
| 8237 | USP11, UHX1 | ubiquitin specific peptidase 11 (EC:3.4.19.12) |
C00044491
|
| 10869 | USP19, ZMYND9 | ubiquitin specific peptidase 19 (EC:3.4.19.12) |
C00044491
|
| 23358 | USP24 | ubiquitin specific peptidase 24 (EC:3.4.19.12) |
C00044491
|
| 84669 | USP32, NY-REN-60, USP10 | ubiquitin specific peptidase 32 (EC:3.4.19.12) |
C00044491
|
| 7375 | USP4, UNP, Unph | ubiquitin specific peptidase 4 (proto-oncogene) (EC:3.4.19.12) |
C00044491
|
| 55230 | USP40 | ubiquitin specific peptidase 40 (EC:3.4.19.12) |
C00044491
|
| 84132 | USP42 | ubiquitin specific peptidase 42 (EC:3.4.19.12) |
C00044491
|
| 84196 | USP48, RAP1GA1, USP31 | ubiquitin specific peptidase 48 (EC:3.4.19.12) |
C00044491
|
| 8239 | USP9X, DFFRX, FAF, FAM | ubiquitin specific peptidase 9, X-linked (EC:3.4.19.12) |
C00044491
|
| 10208 | USPL1, C13orf22, D13S106E, RP11-121O19.1, bA121O19.1 | ubiquitin specific peptidase like 1 |
C00044491
|
| 51118 | UTP11L, CGI94 | UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
C00044491
|
| 9724 | UTP14C, 2700066J21Rik, KIAA0266, UTP14B | UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
C00044491
|
| 51096 | UTP18, CGI-48, WDR50 | UTP18 small subunit (SSU) processome component homolog (yeast) |
C00044491
|
| 57050 | UTP3, CRL1, CRLZ1, SAS10 | UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
C00044491
|
| 7402 | UTRN, DMDL, DRP, DRP1 | utrophin |
C00044491
|
| 57654 | UVSSA, KIAA1530, UVSS3 | UV-stimulated scaffold protein A |
C00044491
|
| 6843 | VAMP1, SYB1, VAMP-1 | vesicle-associated membrane protein 1 (synaptobrevin 1) |
C00044491
|
| 10791 | VAMP5 | vesicle-associated membrane protein 5 |
C00044491
|
| 8673 | VAMP8, EDB, VAMP-8 | vesicle-associated membrane protein 8 |
C00044491
|
| 7407 | VARS, G7A, VARS1, VARS2 | valyl-tRNA synthetase (EC:6.1.1.9) |
C00044491
|
| 57176 | VARS2, G7a, VARS2L, VARSL | valyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.9) |
C00044491
|
| 114990 | VASN, SLITL2 | vasorin |
C00044491
|
| 1462 | VCAN, CSPG2, ERVR, GHAP, PG-M, WGN, WGN1 | versican |
C00044491
|
| 7414 | VCL, CMD1W, CMH15, MV, MVCL | vinculin |
C00044491
|
| 79609 | VCPKMT, C14orf138, METTL21D, VCP-KMT | valosin containing protein lysine (K) methyltransferase |
C00044491
|
| 7419 | VDAC3, HD-VDAC3, VDAC-3 | voltage-dependent anion channel 3 |
C00044491
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00044491
|
| 7716 | VEZF1, DB1, ZNF161 | vascular endothelial zinc finger 1 |
C00044491
|
| 7429 | VIL1, D2S1471, VIL | villin 1 |
C00044491
|
| 81671 | VMP1, EPG3, TANGO5, TMEM49 | vacuole membrane protein 1 |
C00044491
|
| 8876 | VNN1, HDLCQ8, Tiff66 | vanin 1 (EC:3.5.1.92) |
C00044491
|
| 8875 | VNN2, FOAP-4, GPI-80 | vanin 2 (EC:3.5.1.92) |
C00044491
|
| 23230 | VPS13A, CHAC, CHOREIN | vacuolar protein sorting 13 homolog A (S. cerevisiae) |
C00044491
|
| 157680 | VPS13B, CHS1, COH1 | vacuolar protein sorting 13 homolog B (yeast) |
C00044491
|
| 55187 | VPS13D | vacuolar protein sorting 13 homolog D (S. cerevisiae) |
C00044491
|
| 57617 | VPS18, PEP3 | vacuolar protein sorting 18 homolog (S. cerevisiae) |
C00044491
|
| 55737 | VPS35, MEM3, PARK17 | vacuolar protein sorting 35 homolog (S. cerevisiae) |
C00044491
|
| 51028 | VPS36, C13orf9, EAP45 | vacuolar protein sorting 36 homolog (S. cerevisiae) |
C00044491
|
| 79720 | VPS37B | vacuolar protein sorting 37 homolog B (S. cerevisiae) |
C00044491
|
| 23339 | VPS39, TLP, VAM6, hVam6p | vacuolar protein sorting 39 homolog (S. cerevisiae) |
C00044491
|
| 11311 | VPS45, H1, H1VPS45, SCN5, VPS45A, VPS45B, VPS54A, VSP45, VSP45A | vacuolar protein sorting 45 homolog (S. cerevisiae) |
C00044491
|
| 51542 | VPS54, HCC8, SLP-8p, VPS54L, WR, hVps54L | vacuolar protein sorting 54 homolog (S. cerevisiae) |
C00044491
|
| 7443 | VRK1, PCH1, PCH1A | vaccinia related kinase 1 (EC:2.7.11.1) |
C00044491
|
| 51231 | VRK3 | vaccinia related kinase 3 (EC:2.7.11.1) |
C00044491
|
| 54621 | VSIG10 | V-set and immunoglobulin domain containing 10 |
C00044491
|
| 7447 | VSNL1, HLP3, HPCAL3, HUVISL1, VILIP, VILIP-1 | visinin-like 1 |
C00044491
|
| 30813 | VSX1, CAASDS, KTCN, KTCN1, PPCD, PPD, RINX | visual system homeobox 1 |
C00044491
|
| 23078 | VWA8, KIAA0564 | von Willebrand factor A domain containing 8 |
C00044491
|
| 7450 | VWF, F8VWF, VWD | von Willebrand factor |
C00044491
|
| 23063 | WAPAL, FOE, KIAA0261, WAPL | wings apart-like homolog (Drosophila) |
C00044491
|
| 23559 | WBP1, WBP-1 | WW domain binding protein 1 |
C00044491
|
| 54838 | WBP1L, C10orf26, OPA1L, OPAL1 | WW domain binding protein 1-like |
C00044491
|
| 23558 | WBP2, WBP-2 | WW domain binding protein 2 |
C00044491
|
| 155368 | WBSCR27 | Williams Beuren syndrome chromosome region 27 |
C00044491
|
| 11169 | WDHD1, AND-1, CHTF4, CTF4 | WD repeat and HMG-box DNA binding protein 1 |
C00044491
|
| 55717 | WDR11, BRWD2, DR11, HH14, WDR15 | WD repeat domain 11 |
C00044491
|
| 55759 | WDR12, YTM1 | WD repeat domain 12 |
C00044491
|
| 64743 | WDR13, MG21 | WD repeat domain 13 |
C00044491
|
| 57728 | WDR19, ATD5, CED4, DYF-2, IFT144, NPHP13, ORF26, Oseg6, PWDMP | WD repeat domain 19 |
C00044491
|
| 10885 | WDR3, DIP2, UTP12 | WD repeat domain 3 |
C00044491
|
| 114987 | WDR31 | WD repeat domain 31 |
C00044491
|
| 405882 |
C00044491
|
||
| 134430 | WDR36, GLC1G, TA-WDRP, TAWDRP, UTP21 | WD repeat domain 36 |
C00044491
|
| 10785 | WDR4, TRM82, TRMT82 | WD repeat domain 4 |
C00044491
|
| 23160 | WDR43, NET12, UTP5 | WD repeat domain 43 |
C00044491
|
| 22911 | WDR47 | WD repeat domain 47 |
C00044491
|
| 11180 | WDR6 | WD repeat domain 6 |
C00044491
|
| 284403 | WDR62, C19orf14, MCPH2 | WD repeat domain 62 |
C00044491
|
| 23335 | WDR7, TRAG | WD repeat domain 7 |
C00044491
|
| 256764 | WDR72, AI2A3 | WD repeat domain 72 |
C00044491
|
| 54663 | WDR74 | WD repeat domain 74 |
C00044491
|
| 124997 | WDR81, CAMRQ2 | WD repeat domain 81 |
C00044491
|
| 23038 | WDTC1, ADP, DCAF9 | WD and tetratricopeptide repeats 1 |
C00044491
|
| 7465 | WEE1, WEE1A, WEE1hu | WEE1 G2 checkpoint kinase (EC:2.7.10.2) |
C00044491
|
| 7468 | WHSC1, MMSET, NSD2, REIIBP, TRX5, WHS | Wolf-Hirschhorn syndrome candidate 1 (EC:2.1.1.43) |
C00044491
|
| 54904 | WHSC1L1, NSD3, pp14328 | Wolf-Hirschhorn syndrome candidate 1-like 1 (EC:2.1.1.43) |
C00044491
|
| 55062 | WIPI1, ATG18, ATG18A, WIPI49 | WD repeat domain, phosphoinositide interacting 1 |
C00044491
|
| 26100 | WIPI2, ATG18B, Atg21, WIPI-2 | WD repeat domain, phosphoinositide interacting 2 |
C00044491
|
| 7473 | WNT3, INT4 | wingless-type MMTV integration site family, member 3 |
C00044491
|
| 7475 | WNT6 | wingless-type MMTV integration site family, member 6 |
C00044491
|
| 26118 | WSB1, SWIP1, WSB-1 | WD repeat and SOCS box containing 1 |
C00044491
|
| 55884 | WSB2, SBA2 | WD repeat and SOCS box containing 2 |
C00044491
|
| 23286 | WWC1, HBEBP3, HBEBP36, KIBRA | WW and C2 domain containing 1 |
C00044491
|
| 55841 | WWC3, BM042 | WWC family member 3 |
C00044491
|
| 11059 | WWP1, AIP5, Tiul1, hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 |
C00044491
|
| 11060 | WWP2, AIP2, WWp2-like | WW domain containing E3 ubiquitin protein ligase 2 |
C00044491
|
| 25937 | WWTR1, TAZ | WW domain containing transcription regulator 1 |
C00044491
|
| 54739 | XAF1, BIRC4BP, HSXIAPAF1, XIAPAF1 | XIAP associated factor 1 |
C00044491
|
| 7494 | XBP1, TREB5, XBP-1, XBP2 | X-box binding protein 1 |
C00044491
|
| 7498 | XDH, XO, XOR | xanthine dehydrogenase (EC:1.17.1.4 1.17.3.2) |
C00044491
|
| 331 | XIAP, API3, BIRC4, IAP-3, ILP1, MIHA, XLP2, hIAP-3, hIAP3 | X-linked inhibitor of apoptosis |
C00044491
|
| 7508 | XPC, RAD4, XP3, XPCC | xeroderma pigmentosum, complementation group C |
C00044491
|
| 7511 | XPNPEP1, APP1, SAMP, XPNPEP, XPNPEPL, XPNPEPL1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble (EC:3.4.11.9) |
C00044491
|
| 7512 | XPNPEP2, APP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (EC:3.4.11.9) |
C00044491
|
| 23039 | XPO7, EXP7, RANBP16 | exportin 7 |
C00044491
|
| 11260 | XPOT, XPO3 | exportin, tRNA |
C00044491
|
| 9213 | XPR1, SYG1, X3 | xenotropic and polytropic retrovirus receptor 1 |
C00044491
|
| 7516 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 |
C00044491
|
| 7520 | XRCC5, KARP-1, KARP1, KU80, KUB2, Ku86, NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
C00044491
|
| 54464 | XRN1, SEP1 | 5'-3' exoribonuclease 1 |
C00044491
|
| 9942 | XYLB | xylulokinase homolog (H. influenzae) (EC:2.7.1.17) |
C00044491
|
| 57002 | YAE1D1, C7orf36 | Yae1 domain containing 1 |
C00044491
|
| 10413 | YAP1, YAP, YAP2, YAP65, YKI | Yes-associated protein 1 |
C00044491
|
| 54059 | YBEY, C21orf57 | ybeY metallopeptidase (putative) |
C00044491
|
| 646531 |
C00044491
|
||
| 10652 | YKT6 | YKT6 v-SNARE homolog (S. cerevisiae) |
C00044491
|
| 83719 | YPEL3 | yippee-like 3 (Drosophila) |
C00044491
|
| 51646 | YPEL5 | yippee-like 5 (Drosophila) |
C00044491
|
| 64848 | YTHDC2, CAHL | YTH domain containing 2 (EC:3.6.4.13) |
C00044491
|
| 54915 | YTHDF1, C20orf21 | YTH domain family, member 1 |
C00044491
|
| 7532 | YWHAG, 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
C00044491
|
| 284273 | ZADH2 | zinc binding alcohol dehydrogenase domain containing 2 |
C00044491
|
| 51776 | ZAK, AZK, MLK7, MLT, MLTK, MRK, mlklak, pk | sterile alpha motif and leucine zipper containing kinase AZK (EC:2.7.11.25) |
C00044491
|
| 9189 | ZBED1, ALTE, DREF, TRAMP, hDREF | zinc finger, BED-type containing 1 |
C00044491
|
| 58486 | ZBED5, Buster1 | zinc finger, BED-type containing 5 |
C00044491
|
| 113763 | ZBED6CL, C7orf29 | ZBED6 C-terminal like |
C00044491
|
| 27107 | ZBTB11, ZNF-U69274, ZNF913 | zinc finger and BTB domain containing 11 |
C00044491
|
| 7704 | ZBTB16, PLZF, ZNF145 | zinc finger and BTB domain containing 16 |
C00044491
|
| 26137 | ZBTB20, DPZF, HOF, ODA-8S, ZNF288 | zinc finger and BTB domain containing 20 |
C00044491
|
| 49854 | ZBTB21, ZNF295 | zinc finger and BTB domain containing 21 |
C00044491
|
| 7597 | ZBTB25, C14orf51, KUP, ZNF46 | zinc finger and BTB domain containing 25 |
C00044491
|
| 10009 | ZBTB33, ZNF-kaiso, ZNF348 | zinc finger and BTB domain containing 33 |
C00044491
|
| 403341 | ZBTB34, RP11-106H5.1, ZNF918 | zinc finger and BTB domain containing 34 |
C00044491
|
| 253461 | ZBTB38, CIBZ, ZNF921 | zinc finger and BTB domain containing 38 |
C00044491
|
| 23099 | ZBTB43, ZBTB22B, ZNF-X, ZNF297B | zinc finger and BTB domain containing 43 |
C00044491
|
| 29068 | ZBTB44, BTBD15, HSPC063, ZNF851 | zinc finger and BTB domain containing 44 |
C00044491
|
| 51341 | ZBTB7A, FBI-1, FBI1, LRF, ZBTB7, ZNF857A, pokemon | zinc finger and BTB domain containing 7A |
C00044491
|
| 51101 | ZC2HC1A, C8orf70, FAM164A | zinc finger, C2HC-type containing 1A |
C00044491
|
| 23211 | ZC3H4, C19orf7 | zinc finger CCCH-type containing 4 |
C00044491
|
| 23264 | ZC3H7B, RoXaN | zinc finger CCCH-type containing 7B |
C00044491
|
| 92092 | ZC3HAV1L, C7orf39 | zinc finger CCCH-type, antiviral 1-like |
C00044491
|
| 51530 | ZC3HC1, NIPA | zinc finger, C3HC-type containing 1 |
C00044491
|
| 54877 | ZCCHC2, C18orf49 | zinc finger, CCHC domain containing 2 |
C00044491
|
| 219654 | ZCCHC24, C10orf56 | zinc finger, CCHC domain containing 24 |
C00044491
|
| 79670 | ZCCHC6, PAPD6 | zinc finger, CCHC domain containing 6 (EC:2.7.7.52) |
C00044491
|
| 55596 | ZCCHC8 | zinc finger, CCHC domain containing 8 |
C00044491
|
| 79683 | ZDHHC14, NEW1CP | zinc finger, DHHC-type containing 14 (EC:2.3.1.225) |
C00044491
|
| 84287 | ZDHHC16, APH2 | zinc finger, DHHC-type containing 16 (EC:2.3.1.225) |
C00044491
|
| 84243 | ZDHHC18, DHHC-18, DHHC18 | zinc finger, DHHC-type containing 18 (EC:2.3.1.225) |
C00044491
|
| 25921 | ZDHHC5, DHHC5, ZNF375 | zinc finger, DHHC-type containing 5 (EC:2.3.1.225) |
C00044491
|
| 64429 | ZDHHC6, ZNF376 | zinc finger, DHHC-type containing 6 (EC:2.3.1.225) |
C00044491
|
| 6935 | ZEB1, AREB6, BZP, DELTAEF1, FECD6, NIL2A, PPCD3, TCF8, ZFHEP, ZFHX1A | zinc finger E-box binding homeobox 1 |
C00044491
|
| 90637 | ZFAND2A, AIRAP | zinc finger, AN1-type domain 2A |
C00044491
|
| 7763 | ZFAND5, ZA20D2, ZFAND5A, ZNF216 | zinc finger, AN1-type domain 5 |
C00044491
|
| 54469 | ZFAND6, AWP1, ZA20D3, ZFAND5B | zinc finger, AN1-type domain 6 |
C00044491
|
| 441951 | ZFAS1, C20orf199, HSUP1, HSUP2, NCRNA00275, ZNFX1-AS1 | ZNFX1 antisense RNA 1 |
C00044491
|
| 677 | ZFP36L1, BRF1, Berg36, ERF-1, ERF1, RNF162B, TIS11B, cMG1 | ZFP36 ring finger protein-like 1 |
C00044491
|
| 678 | ZFP36L2, BRF2, ERF-2, ERF2, RNF162C, TIS11D | ZFP36 ring finger protein-like 2 |
C00044491
|
| 139735 | ZFP92, ZNF897 | ZFP92 zinc finger protein |
C00044491
|
| 161882 | ZFPM1, FOG, FOG1, ZC2HC11A, ZNF408, ZNF89A | zinc finger protein, FOG family member 1 |
C00044491
|
| 23414 | ZFPM2, DIH3, FOG2, ZC2HC11B, ZNF89B, hFOG-2 | zinc finger protein, FOG family member 2 |
C00044491
|
| 53349 | ZFYVE1, DFCP1, SR3, TAFF1, ZNFN2A1 | zinc finger, FYVE domain containing 1 |
C00044491
|
| 9765 | ZFYVE16, PPP1R69 | zinc finger, FYVE domain containing 16 |
C00044491
|
| 84936 | ZFYVE19, MPFYVE | zinc finger, FYVE domain containing 19 |
C00044491
|
| 22882 | ZHX2, AFR1, RAF | zinc fingers and homeoboxes 2 |
C00044491
|
| 64393 | ZMAT3, PAG608, WIG-1, WIG1 | zinc finger, matrin-type 3 |
C00044491
|
| 79830 | ZMYM1, MYM, RP11-181E22.4 | zinc finger, MYM-type 1 |
C00044491
|
| 9202 | ZMYM4, CDIR, MYM, ZNF198L3, ZNF262 | zinc finger, MYM-type 4 |
C00044491
|
| 23613 | ZMYND8, PRKCBP1, PRO2893, RACK7 | zinc finger, MYND-type containing 8 |
C00044491
|
| 163227 | ZNF100 | zinc finger protein 100 |
C00044491
|
| 64397 | ZNF106, SH3BP3, ZFP106, ZNF474 | zinc finger protein 106 |
C00044491
|
| 7559 | ZNF12, GIOT-3, HZF11, KOX3, ZNF325 | zinc finger protein 12 |
C00044491
|
| 7705 | ZNF146, OZF | zinc finger protein 146 |
C00044491
|
| 7743 | ZNF189 | zinc finger protein 189 |
C00044491
|
| 7752 | ZNF200 | zinc finger protein 200 |
C00044491
|
| 7756 | ZNF207 | zinc finger protein 207 |
C00044491
|
| 7570 | ZNF22, HKR-T1, KOX15, ZNF422, Zfp422 | zinc finger protein 22 |
C00044491
|
| 7772 | ZNF229 | zinc finger protein 229 |
C00044491
|
| 10781 | ZNF266, HZF1 | zinc finger protein 266 |
C00044491
|
| 10308 | ZNF267, HZF2 | zinc finger protein 267 |
C00044491
|
| 10793 | ZNF273, HZF9 | zinc finger protein 273 |
C00044491
|
| 140883 | ZNF280B, 5'OY11.1, D87009.C22.3, SUHW2, ZNF279, ZNF632 | zinc finger protein 280B |
C00044491
|
| 55609 | ZNF280C, SUHW3, ZNF633 | zinc finger protein 280C |
C00044491
|
| 23528 | ZNF281, ZBP-99, ZNP-99 | zinc finger protein 281 |
C00044491
|
| 55900 | ZNF302, HSD16, MST154, MSTP154, ZNF135L, ZNF140L, ZNF327 | zinc finger protein 302 |
C00044491
|
| 26152 | ZNF337 | zinc finger protein 337 |
C00044491
|
| 7581 | ZNF33A, KOX2, KOX31, KOX5, NF11A, ZNF11, ZNF11A, ZNF33, ZZAPK | zinc finger protein 33A |
C00044491
|
| 149076 | ZNF362, RN, lin-29 | zinc finger protein 362 |
C00044491
|
| 195828 | ZNF367, AFF29, CDC14B, ZFF29 | zinc finger protein 367 |
C00044491
|
| 84911 | ZNF382, KS1 | zinc finger protein 382 |
C00044491
|
| 346157 | ZNF391, dJ153G14.3 | zinc finger protein 391 |
C00044491
|
| 55786 | ZNF415 | zinc finger protein 415 |
C00044491
|
| 126299 | ZNF428, C19orf37, Zfp428 | zinc finger protein 428 |
C00044491
|
| 80264 | ZNF430 | zinc finger protein 430 |
C00044491
|
| 170959 | ZNF431 | zinc finger protein 431 |
C00044491
|
| 126070 | ZNF440 | zinc finger protein 440 |
C00044491
|
| 10224 | ZNF443, ZK1 | zinc finger protein 443 |
C00044491
|
| 55311 | ZNF444, EZF-2, EZF2, ZSCAN17 | zinc finger protein 444 |
C00044491
|
| 353274 | ZNF445, ZKSCAN15, ZNF168, ZSCAN47 | zinc finger protein 445 |
C00044491
|
| 84627 | ZNF469, BCS | zinc finger protein 469 |
C00044491
|
| 25888 | ZNF473, HZFP100, ZN473 | zinc finger protein 473 |
C00044491
|
| 197407 | ZNF48, ZNF553 | zinc finger protein 48 |
C00044491
|
| 158399 | ZNF483, ZKSCAN16, ZSCAN48 | zinc finger protein 483 |
C00044491
|
| 84838 | ZNF496, NIZP1, ZFP496, ZKSCAN17, ZSCAN49 | zinc finger protein 496 |
C00044491
|
| 100131213 | ZNF503-AS2, C10orf41, NCRNA00245 | ZNF503 antisense RNA 2 |
C00044491
|
| 22847 | ZNF507 | zinc finger protein 507 |
C00044491
|
| 57473 | ZNF512B, GM632 | zinc finger protein 512B |
C00044491
|
| 162655 | ZNF519, HsT2362 | zinc finger protein 519 |
C00044491
|
| 163255 | ZNF540, Nbla10512 | zinc finger protein 540 |
C00044491
|
| 162972 | ZNF550 | zinc finger protein 550 |
C00044491
|
| 148254 | ZNF555 | zinc finger protein 555 |
C00044491
|
| 93134 | ZNF561 | zinc finger protein 561 |
C00044491
|
| 126295 | ZNF57, ZNF424 | zinc finger protein 57 |
C00044491
|
| 90317 | ZNF616 | zinc finger protein 616 |
C00044491
|
| 199692 | ZNF627 | zinc finger protein 627 |
C00044491
|
| 27332 | ZNF638, NP220, ZFML, Zfp638 | zinc finger protein 638 |
C00044491
|
| 22834 | ZNF652 | zinc finger protein 652 |
C00044491
|
| 79027 | ZNF655, VIK, VIK-1 | zinc finger protein 655 |
C00044491
|
| 7617 |
C00044491
|
||
| 340252 | ZNF680 | zinc finger protein 680 |
C00044491
|
| 115509 | ZNF689, TIPUH1 | zinc finger protein 689 |
C00044491
|
| 57116 | ZNF695, RP11-551G24.1, SBZF3 | zinc finger protein 695 |
C00044491
|
| 90874 | ZNF697 | zinc finger protein 697 |
C00044491
|
| 440519 | ZNF724P | zinc finger protein 724, pseudogene |
C00044491
|
| 155061 | ZNF746, PARIS | zinc finger protein 746 |
C00044491
|
| 54989 | ZNF770, PRO1914 | zinc finger protein 770 |
C00044491
|
| 7633 | ZNF79, pT7 | zinc finger protein 79 |
C00044491
|
| 390980 | ZNF805 | zinc finger protein 805 |
C00044491
|
| 152485 | ZNF827 | zinc finger protein 827 |
C00044491
|
| 55769 | ZNF83, HPF1, ZNF816B | zinc finger protein 83 |
C00044491
|
| 401898 | ZNF833P, ZNF833 | zinc finger protein 833, pseudogene |
C00044491
|
| 284391 | ZNF844 | zinc finger protein 844 |
C00044491
|
| 7643 | ZNF90, HTF9 | zinc finger protein 90 |
C00044491
|
| 54680 | ZNHIT6, BCD1, C1orf181, NY-BR-75 | zinc finger, HIT-type containing 6 |
C00044491
|
| 84133 | ZNRF3, BK747E2.3, RNF203 | zinc and ring finger 3 |
C00044491
|
| 222696 | ZSCAN23, ZNF390, ZNF453, dJ29K1.3, dJ29K1.3.1 | zinc finger and SCAN domain containing 23 |
C00044491
|
| 201516 | ZSCAN4, ZNF494 | zinc finger and SCAN domain containing 4 |
C00044491
|
| 90204 | ZSWIM1, C20orf162 | zinc finger, SWIM-type containing 1 |
C00044491
|
| 57688 | ZSWIM6 | zinc finger, SWIM-type containing 6 |
C00044491
|
| 11130 | ZWINT, HZwint-1, KNTC2AP, ZWINT1 | ZW10 interacting kinetochore protein |
C00044491
|
| 7789 | ZXDA, ZNF896 | zinc finger, X-linked, duplicated A |
C00044491
|
| 79364 | ZXDC, ZXDL | ZXD family zinc finger C |
C00044491
|
| 79699 | ZYG11B, ZYG11 | zyg-11 family member B, cell cycle regulator |
C00044491
|
| 23140 | ZZEF1, ZZZ4 | zinc finger, ZZ-type with EF-hand domain 1 |
C00044491
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (60)
| OMIM | preferred title | UniProt |
|---|---|---|
| #100100 | Abdominal muscles, absence of, with urinary tract abnormality and cryptorchidism |
P20309
|
| #103780 | Alcohol dependence |
P08172
P14416 P31645 |
| #614373 | Amyotrophic lateral sclerosis 16, juvenile; als16 |
Q99720
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #602025 | Body mass index quantitative trait locus 9; bmiq9 |
P41968
|
| #300615 | Brunner syndrome |
P21397
|
| #114500 | Colorectal cancer; crc |
P84022
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #162800 | Cyclic neutropenia |
P08246
|
| #612522 | Diabetes mellitus, insulin-dependent, 22; iddm22 |
P51681
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613659 | Gastric cancer |
P04626
|
| #137215 | Gastric cancer, hereditary diffuse; hdgc |
P04626
|
| #231095 | Ghosal hematodiaphyseal dysplasia; ghdd |
P24557
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137800 | Glioma susceptibility 1; glm1 |
P04626
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #609423 | Human immunodeficiency virus type 1, susceptibility to |
P41597
P51681 |
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #211980 | Lung cancer |
P00533
P04626 |
| #608516 | Major depressive disorder; mdd |
P08172
|
| %300852 | Mental retardation, x-linked 88; mrx88 |
P50052
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #126200 | Multiple sclerosis, susceptibility to; ms |
P08575
|
| #159900 | Myoclonic dystonia |
P14416
|
| #202700 | Neutropenia, severe congenital, 1, autosomal dominant; scn1 |
P08246
|
| #601665 | Obesity |
P32245
|
| #164230 | Obsessive-compulsive disorder; ocd |
P31645
|
| #604715 | Orthostatic intolerance |
P23975
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #167000 | Ovarian cancer |
P04626
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #613135 | Parkinsonism-dystonia, infantile; pkdys |
Q01959
|
| #172700 | Pick disease of brain |
P10636
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #608971 | Severe combined immunodeficiency, autosomal recessive, t cell-negative, b cell-positive, nk cell-positive |
P08575
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #601367 | Stroke, ischemic |
P20292
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #190300 | Tremor, hereditary essential, 1; etm1 |
P35462
|
| #610379 | West nile virus, susceptibility to |
P51681
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (51)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04626 (related) |
| H00022 | Bladder cancer |
P00533
(related)
P04626 (related) |
| H00028 | Choriocarcinoma |
P00533
(related)
P03956 (related) P04626 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P04626 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04626 (related) Q92731 (marker) |
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
|
| H00019 | Pancreatic cancer |
P04626
(related)
|
| H00027 | Ovarian cancer |
P04626
(related)
|
| H00031 | Breast cancer |
P04626
(related)
P04626 (marker) |
| H00046 | Cholangiocarcinoma |
P04626
(related)
P35354 (related) |
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H00100 | Neutropenic disorders |
P08246
(related)
|
| H00091 | T-B+Severe combined immunodeficiencies (SCIDs) |
P08575
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00025 | Penile cancer |
P14780
(related)
P35354 (related) |
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H01031 | Orthostatic intolerance (OI) |
P23975
(related)
|
| H00490 | Diaphyseal dysplasia with anemia (Ghosal) |
P24557
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
P50052
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q03164
(related)
Q03164 (marker) |
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|
Diseases related to CTD interactions
166 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D015746 | Abdominal Pain |
C00044491
|
| D000014 | Abnormalities, Drug-Induced |
C00044491
|
| D009008 | Abnormalities, Severe Teratoid |
C00044491
|
| D000138 | Acidosis |
C00044491
|
| D000140 | Acidosis, Lactic |
C00044491
|
| D058186 | Acute Kidney Injury |
C00044491
|
| D000402 | Airway Obstruction |
C00044491
|
| D020751 | Alcohol-Induced Disorders |
C00044491
|
| D000707 | Anaphylaxis |
C00044491
|
| D000740 | Anemia |
C00044491
|
| D000743 | Anemia, Hemolytic |
C00044491
|
| D000799 | Angioedema |
C00044491
|
| D000855 | Anorexia |
C00044491
|
| D000860 | Anoxia |
C00044491
|
| D001002 | Anuria |
C00044491
|
| D001161 | Arteriosclerosis |
C00044491
|
| D018771 | Arthralgia |
C00044491
|
| D015210 | Arthritis, Gouty |
C00044491
|
| D001172 | Arthritis, Rheumatoid |
C00044491
|
| D001249 | Asthma |
C00044491
|
| D001281 | Atrial Fibrillation |
C00044491
|
| D001321 | Autistic Disorder |
C00044491
|
| D053099 | Azotemia |
C00044491
|
| D001649 | Bile Duct Diseases |
C00044491
|
| D001660 | Biliary Tract Diseases |
C00044491
|
| D019575 | Blindness, Cortical |
C00044491
|
| D001919 | Bradycardia |
C00044491
|
| D001927 | Brain Diseases |
C00044491
|
| D001929 | Brain Edema |
C00044491
|
| D006528 | Carcinoma, Hepatocellular |
C00044491
|
| D002292 | Carcinoma, Renal Cell |
C00044491
|
| D016534 | Cardiac Output, High |
C00044491
|
| D009202 | Cardiomyopathies |
C00044491
|
| D002318 | Cardiovascular Diseases |
C00044491
|
| D002375 | Catalepsy |
C00044491
|
| D002386 | Cataract |
C00044491
|
| D002779 | Cholestasis |
C00044491
|
| D015267 | Churg-Strauss Syndrome |
C00044491
|
| D003110 | Colonic Neoplasms |
C00044491
|
| D003128 | COMA |
C00044491
|
| D003221 | Confusion |
C00044491
|
| D003424 | Crohn Disease |
C00044491
|
| D003645 | Death, Sudden |
C00044491
|
| D004195 | Disease Models, Animal |
C00044491
|
| D003875 | Drug Eruptions |
C00044491
|
| D004342 | Drug Hypersensitivity |
C00044491
|
| D056486 | Drug-Induced Liver Injury |
C00044491
|
| D056487 | Drug-Induced Liver Injury, Chronic |
C00044491
|
| D062787 | Drug Overdose |
C00044491
|
| D064420 | Drug-Related Side Effects and Adverse Reactions |
C00044491
|
| D004830 | Epilepsy, Tonic-Clonic |
C00044491
|
| D005076 | Exanthema |
C00044491
|
| D005094 | Exophthalmos |
C00044491
|
| D005157 | Facial Pain |
C00044491
|
| D005234 | Fatty Liver |
C00044491
|
| D005313 | Fetal Death |
C00044491
|
| D005334 | Fever |
C00044491
|
| D005756 | Gastritis |
C00044491
|
| D006471 | Gastrointestinal Hemorrhage |
C00044491
|
| D006099 | Granuloma |
C00044491
|
| D006212 | Hallucinations |
C00044491
|
| D006261 | Headache |
C00044491
|
| D020773 | Headache Disorders |
C00044491
|
| D006461 | Hemolysis |
C00044491
|
| D006470 | Hemorrhage |
C00044491
|
| D006501 | Hepatic Encephalopathy |
C00044491
|
| D006505 | Hepatitis |
C00044491
|
| D006529 | Hepatomegaly |
C00044491
|
| D006930 | Hyperalgesia |
C00044491
|
| D022124 | Hyperammonemia |
C00044491
|
| D006940 | Hyperemia |
C00044491
|
| D006941 | Hyperesthesia |
C00044491
|
| D006965 | Hyperplasia |
C00044491
|
| D006973 | Hypertension |
C00044491
|
| D006984 | Hypertrophy |
C00044491
|
| D006985 | Hyperventilation |
C00044491
|
| D034141 | Hypoalbuminemia |
C00044491
|
| D007003 | Hypoglycemia |
C00044491
|
| D007008 | Hypokalemia |
C00044491
|
| D017674 | Hypophosphatemia |
C00044491
|
| D007022 | Hypotension |
C00044491
|
| D007035 | Hypothermia |
C00044491
|
| D020896 | Hypovolemia |
C00044491
|
| D007249 | Inflammation |
C00044491
|
| D019586 | Intracranial Hypertension |
C00044491
|
| D007565 | Jaundice |
C00044491
|
| D007674 | Kidney Diseases |
C00044491
|
| D007676 | Kidney Failure, Chronic |
C00044491
|
| D007680 | Kidney Neoplasms |
C00044491
|
| D007681 | Kidney Papillary Necrosis |
C00044491
|
| D007683 | Kidney Tubular Necrosis, Acute |
C00044491
|
| D007859 | Learning Disorders |
C00044491
|
| D053609 | Lethargy |
C00044491
|
| D007970 | Leukopenia |
C00044491
|
| D008103 | Liver Cirrhosis |
C00044491
|
| D008107 | Liver Diseases |
C00044491
|
| D017093 | Liver Failure |
C00044491
|
| D017114 | Liver Failure, Acute |
C00044491
|
| D008113 | Liver Neoplasms |
C00044491
|
| D008114 | Liver Neoplasms, Experimental |
C00044491
|
| D008133 | Long QT Syndrome |
C00044491
|
| D008171 | Lung Diseases |
C00044491
|
| D047508 | Massive Hepatic Necrosis |
C00044491
|
| D008545 | Melanoma |
C00044491
|
| D008546 | Melanoma, Experimental |
C00044491
|
| D001523 | Mental Disorders |
C00044491
|
| D008708 | Methemoglobinemia |
C00044491
|
| D008881 | Migraine Disorders |
C00044491
|
| D018777 | Multiple Chemical Sensitivity |
C00044491
|
| D009102 | Multiple Organ Failure |
C00044491
|
| D015878 | Mydriasis |
C00044491
|
| D009203 | Myocardial Infarction |
C00044491
|
| D009207 | Myoclonus |
C00044491
|
| D009325 | Nausea |
C00044491
|
| D009336 | Necrosis |
C00044491
|
| D009393 | Nephritis |
C00044491
|
| D009395 | Nephritis, Interstitial |
C00044491
|
| D009397 | Nephrocalcinosis |
C00044491
|
| D009436 | Neural Tube Defects |
C00044491
|
| D019954 | Neurobehavioral Manifestations |
C00044491
|
| D019636 | Neurodegenerative Diseases |
C00044491
|
| D009459 | Neuroleptic Malignant Syndrome |
C00044491
|
| D009846 | Oliguria |
C00044491
|
| D010003 | Osteoarthritis |
C00044491
|
| D010146 | Pain |
C00044491
|
| D010149 | Pain, Postoperative |
C00044491
|
| D010195 | Pancreatitis |
C00044491
|
| D010198 | Pancytopenia |
C00044491
|
| D011183 | Postoperative Complications |
C00044491
|
| D020250 | Postoperative Nausea and Vomiting |
C00044491
|
| D011230 | Precancerous Conditions |
C00044491
|
| D011248 | Pregnancy Complications |
C00044491
|
| D011507 | Proteinuria |
C00044491
|
| D011537 | Pruritus |
C00044491
|
| D011595 | Psychomotor Agitation |
C00044491
|
| D011654 | Pulmonary Edema |
C00044491
|
| D011693 | Purpura |
C00044491
|
| D055665 | Purpura Fulminans |
C00044491
|
| D011695 | Purpura, Schoenlein-Henoch |
C00044491
|
| D016553 | Purpura, Thrombocytopenic, Idiopathic |
C00044491
|
| D051437 | Renal Insufficiency |
C00044491
|
| D012130 | Respiratory Hypersensitivity |
C00044491
|
| D012131 | Respiratory Insufficiency |
C00044491
|
| D012141 | Respiratory Tract Infections |
C00044491
|
| D012185 | Retroperitoneal Fibrosis |
C00044491
|
| D012206 | Rhabdomyolysis |
C00044491
|
| D012640 | Seizures |
C00044491
|
| D012852 | Sinusitis |
C00044491
|
| D055009 | Spondylosis |
C00044491
|
| D013226 | Status Epilepticus |
C00044491
|
| D013262 | Stevens-Johnson Syndrome |
C00044491
|
| D013272 | Stomach Diseases |
C00044491
|
| D020521 | Stroke |
C00044491
|
| D013375 | Substance Withdrawal Syndrome |
C00044491
|
| D017180 | Tachycardia, Ventricular |
C00044491
|
| D013927 | Thrombosis |
C00044491
|
| D016171 | Torsades de Pointes |
C00044491
|
| D014516 | Ureteral Neoplasms |
C00044491
|
| D014517 | Ureteral Obstruction |
C00044491
|
| D001749 | Urinary Bladder Neoplasms |
C00044491
|
| D014570 | Urologic Diseases |
C00044491
|
| D014581 | Urticaria |
C00044491
|
| D014652 | Vascular Diseases |
C00044491
|
| D057772 | Vascular System Injuries |
C00044491
|
| D018366 | Vasculitis, Leukocytoclastic, Cutaneous |
C00044491
|
| D014839 | Vomiting |
C00044491
|