Picramnia pentandra
Species
KNApSAcK Entry
| Organism name | Picramnia pentandra |
|---|---|
| Genus | Picramnia |
| Family | Picramniaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Picramnia pentandra |
|---|---|
| Linked NCBI taxonomy ID | 85239 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Picramniaceae |
|---|---|
| ID | 85159 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Natural Activity
List (10)
| Species | Activity |
|---|---|
| Picramnia pentandra Sw. | Antifertility |
| Picramnia pentandra Sw. | Antispasmodic |
| Picramnia pentandra Sw. | Aphrodisiac |
| Picramnia pentandra Sw. | Carminative |
| Picramnia pentandra Sw. | Diuretic |
| Picramnia pentandra Sw. | Febrifuge |
| Picramnia pentandra Sw. | Orexigenic |
| Picramnia pentandra Sw. | Sedative |
| Picramnia pentandra Sw. | Tonic |
| Picramnia pentandra Sw. | Vermifuge |
Metabolite list (1)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00034390
|
3-epi-Betulinic acid
|
CHEMBL269277
CHEMBL71690 CHEMBL519059 CHEMBL1318530 CHEMBL2005635 |
34 / 17 / 14 | No. 23 | No. 51 |
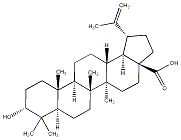
|
Human Protein / Gene in interactions
34 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00034390 | 1 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00034390 | 2 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00034390 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00034390 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00034390 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00034390 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00034390 | 11 / 10 |
| Q02156 | Protein kinase C epsilon type | Eta | C00034390 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00034390 | 0 / 1 |
| P15559 | NAD(P)H dehydrogenase [quinone] 1 | Enzyme | C00034390 | 0 / 0 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00034390 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00034390 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00034390 | 0 / 0 |
| P08151 | Zinc finger protein GLI1 | Unclassified protein | C00034390 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00034390 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00034390 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00034390 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00034390 | 0 / 0 |
| Q04206 | Transcription factor p65 | Transcription Factor | C00034390 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00034390 | 1 / 1 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00034390 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00034390 | 0 / 0 |
| Q13133 | Oxysterols receptor LXR-alpha | NR1H3 | C00034390 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00034390 | 0 / 0 |
| P55055 | Oxysterols receptor LXR-beta | NR1H3 | C00034390 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00034390 | 0 / 0 |
| P05771 | Protein kinase C beta type | Alpha | C00034390 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00034390 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00034390 | 0 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00034390 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00034390 | 1 / 0 |
| Q8IUX4 | DNA dC->dU-editing enzyme APOBEC-3F | Enzyme | C00034390 | 0 / 0 |
| Q9UBT2 | SUMO-activating enzyme subunit 2 | Enzyme | C00034390 | 0 / 0 |
| Q9UBE0 | SUMO-activating enzyme subunit 1 | Unclassified protein | C00034390 | 0 / 0 |
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (17)
| OMIM | preferred title | UniProt |
|---|---|---|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (14)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|