Beta vulgaris L. var. saccharifera
Species
KNApSAcK Entry
| Organism name | Beta vulgaris L. var. saccharifera |
|---|---|
| Genus | Beta |
| Family | Chenopodiaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Amaranthaceae |
|---|---|
| Linked NCBI taxonomy ID | 3563 |
| Linked level | family |
Family
| Family in NCBI taxonomy | Amaranthaceae |
|---|---|
| ID | 3563 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (1)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
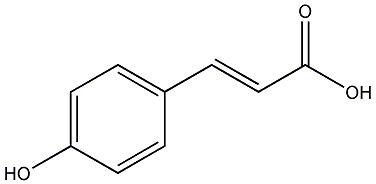
C00000152
|
p-Coumaric acid
|
CHEMBL66879
CHEMBL2336752 |
C032171
|
23 / 13 / 18 | 6 / 1 | No. 904 | No. 6 |
|
Human Protein / Gene in interactions
23 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P17516 | Aldo-keto reductase family 1 member C4 | Enzyme | C00000152 | 1 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00000152 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000152 | 0 / 3 |
| Q13093 | Platelet-activating factor acetylhydrolase | Enzyme | C00000152 | 3 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000152 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000152 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000152 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00000152 | 1 / 2 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000152 | 1 / 8 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000152 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000152 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000152 | 1 / 2 |
| P42330 | Aldo-keto reductase family 1 member C3 | Enzyme | C00000152 | 0 / 0 |
| Q8TDS4 | Hydroxycarboxylic acid receptor 2 | Hydroxycarboxylic acid receptor | C00000152 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00000152 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000152 | 0 / 0 |
| P14679 | Tyrosinase | Oxidoreductase | C00000152 | 4 / 2 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000152 | 0 / 1 |
| Q04828 | Aldo-keto reductase family 1 member C1 | Enzyme | C00000152 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00000152 | 1 / 1 |
| P52895 | Aldo-keto reductase family 1 member C2 | Enzyme | C00000152 | 1 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000152 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000152 | 1 / 1 |
6 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00000152
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00000152
|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00000152
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00000152
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000152
|
| 7299 | TYR, ATN, CMM8, OCA1, OCA1A, OCAIA, SHEP3 | tyrosinase (EC:1.14.18.1) |
C00000152
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (13)
| OMIM | preferred title | UniProt |
|---|---|---|
| #614279 | 46,xy sex reversal 8; srxy8 |
P17516
P52895 |
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #600807 | Asthma, susceptibility to |
Q13093
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #147050 | Ige responsiveness, atopic; iger |
Q13093
|
| #211980 | Lung cancer |
P00533
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #614278 | Platelet-activating factor acetylhydrolase deficiency; pafad |
Q13093
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
KEGG DISEASE (18)
| KEGG | name | UniProt |
|---|---|---|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
|
| H00022 | Bladder cancer |
P00533
(related)
|
| H00028 | Choriocarcinoma |
P00533
(related)
|
| H00030 | Cervical cancer |
P00533
(related)
|
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00038 | Malignant melanoma |
P14679
(marker)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|