Lilium candidum
Species
KNApSAcK Entry
| Organism name | Lilium candidum |
|---|---|
| Genus | Lilium |
| Family | Liliaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Lilium candidum |
|---|---|
| Linked NCBI taxonomy ID | 83802 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Liliaceae |
|---|---|
| ID | 4677 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | Liliopsida |
|---|---|
| ID | 4447 |
Natural Activity
List (13)
| Species | Activity |
|---|---|
| Lilium candidum L. | Analgesic |
| Lilium candidum L. | Antiinflammatory |
| Lilium candidum L. | Antispasmodic |
| Lilium candidum L. | Astringent |
| Lilium candidum L. | Demulcent |
| Lilium candidum L. | Diaphoretic |
| Lilium candidum L. | Diuretic |
| Lilium candidum L. | Emollient |
| Lilium candidum L. | Expectorant |
| Lilium candidum L. | Fungicide |
| Lilium candidum L. | Phagocytotic |
| Lilium candidum L. | Stimulant |
| Lilium candidum L. | Tonic |
Metabolite list (5)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
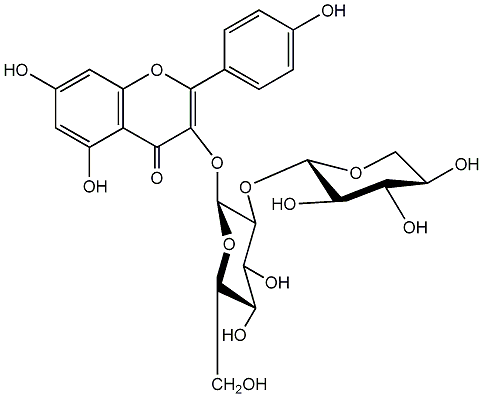
C00005163
|
Leucoside
/ Kaempferol 3-sambubioside / 5,7-Dihydroxy-2-(4-hydroxyphenyl)-3-[(2-O-beta-D-xylopyranosyl-beta-D-glucopyranosyl)oxy]-4H-1-benzopyran-4-one |
CHEMBL446574
CHEMBL1096690 |
No. 1 | No. 15 |
|
|||
|
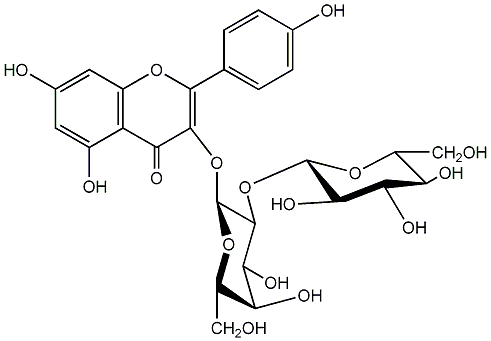
C00005157
|
Panasenoside
|
CHEMBL500233
CHEMBL1375795 CHEMBL2032411 |
12 / 2 / 2 | No. 1 | No. 15 |
|
||
|
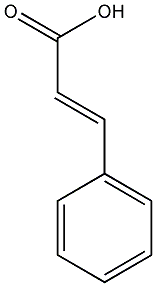
C00000170
|
trans-Cinnamate
/ trans-Cinnamic acid |
CHEMBL27246
|
5 / 2 / 2 | No. 904 | No. 6 |
|
||
|
C00002347
|
Lilaline
|
No. 3419 | No. 15 |

|
||||
|
C00005002
|
3,5,7,4'-Tetrahydroxy-8-C-(3-methylsuccinoyl)flavone
/ 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)-alpha-methyl-gamma,4-dioxo-4H-1-benzopyran-8-butanoic acid |
No. 8029 |

|
Human Protein / Gene in interactions
17 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00000170 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00005157 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00005157 | 1 / 1 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00005157 | 0 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00000170 | 1 / 1 |
| Q8TDS4 | Hydroxycarboxylic acid receptor 2 | Hydroxycarboxylic acid receptor | C00000170 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00005157 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00005157 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00005157 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00000170 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00005157 | 1 / 1 |
| P03372 | Estrogen receptor | NR3A1 | C00000170 | 1 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00005157 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00005157 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00005157 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00005157 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00005157 | 0 / 0 |