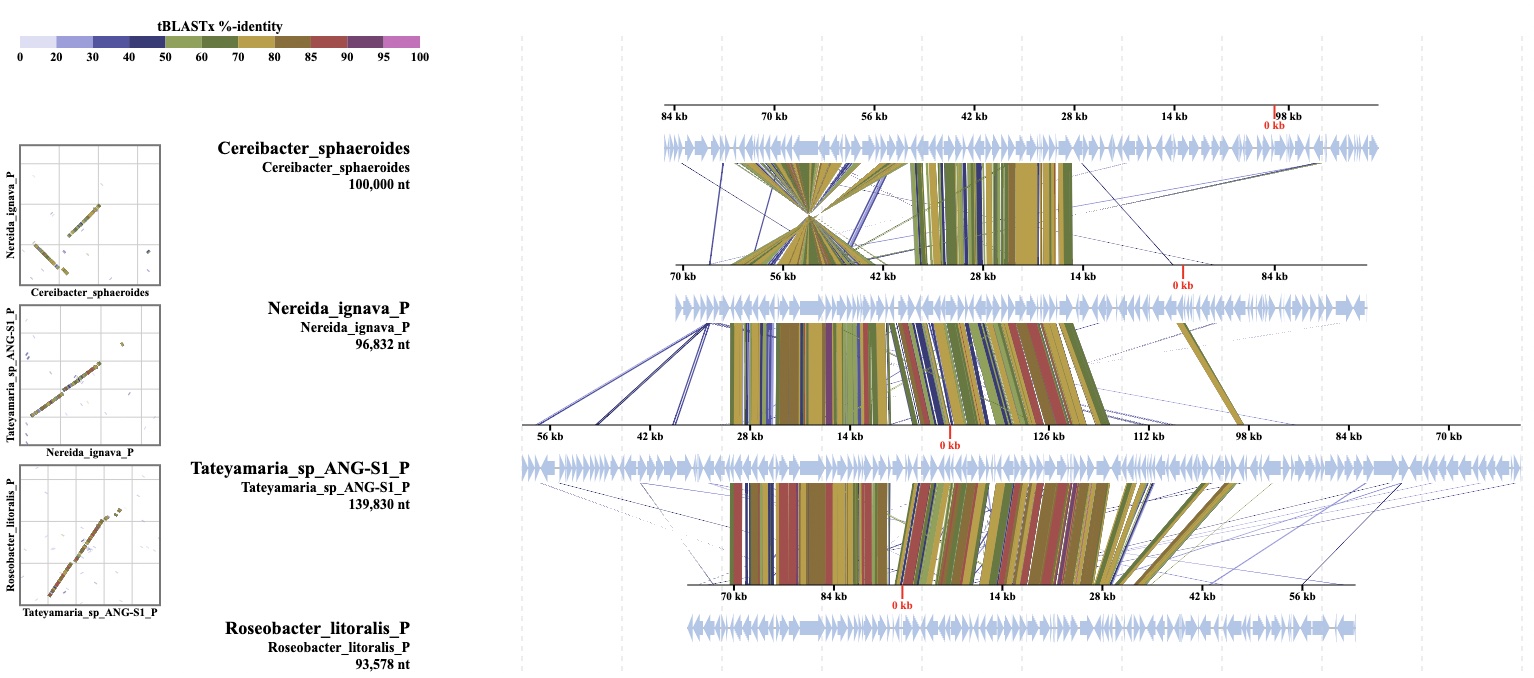
Alignment
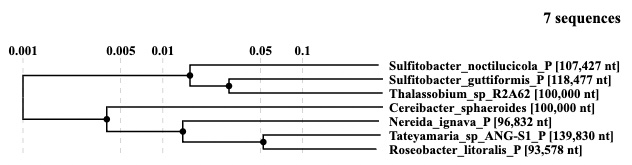
Guide tree
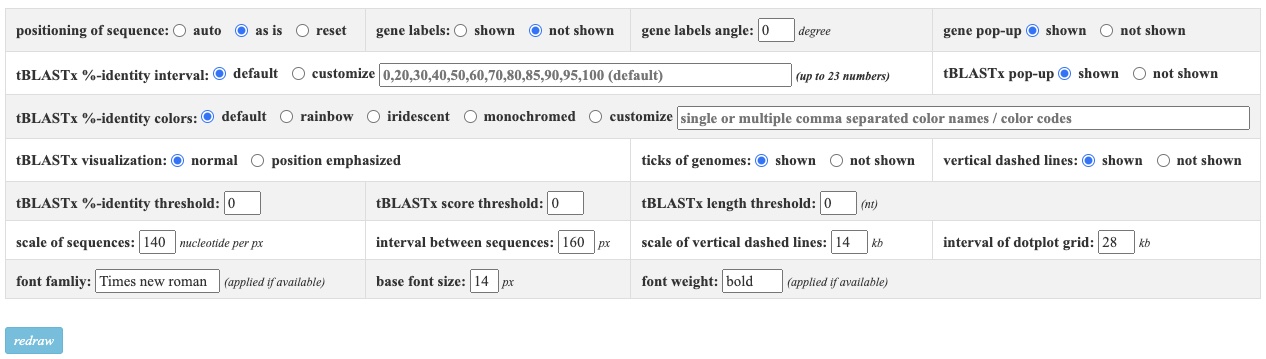
Parameter configuration
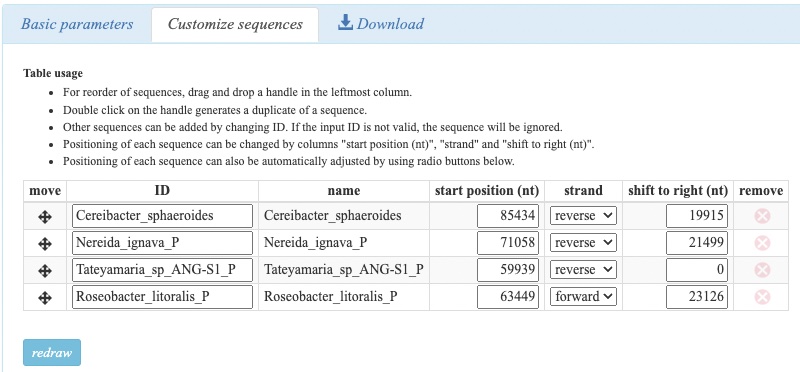
Reordering and position optimization
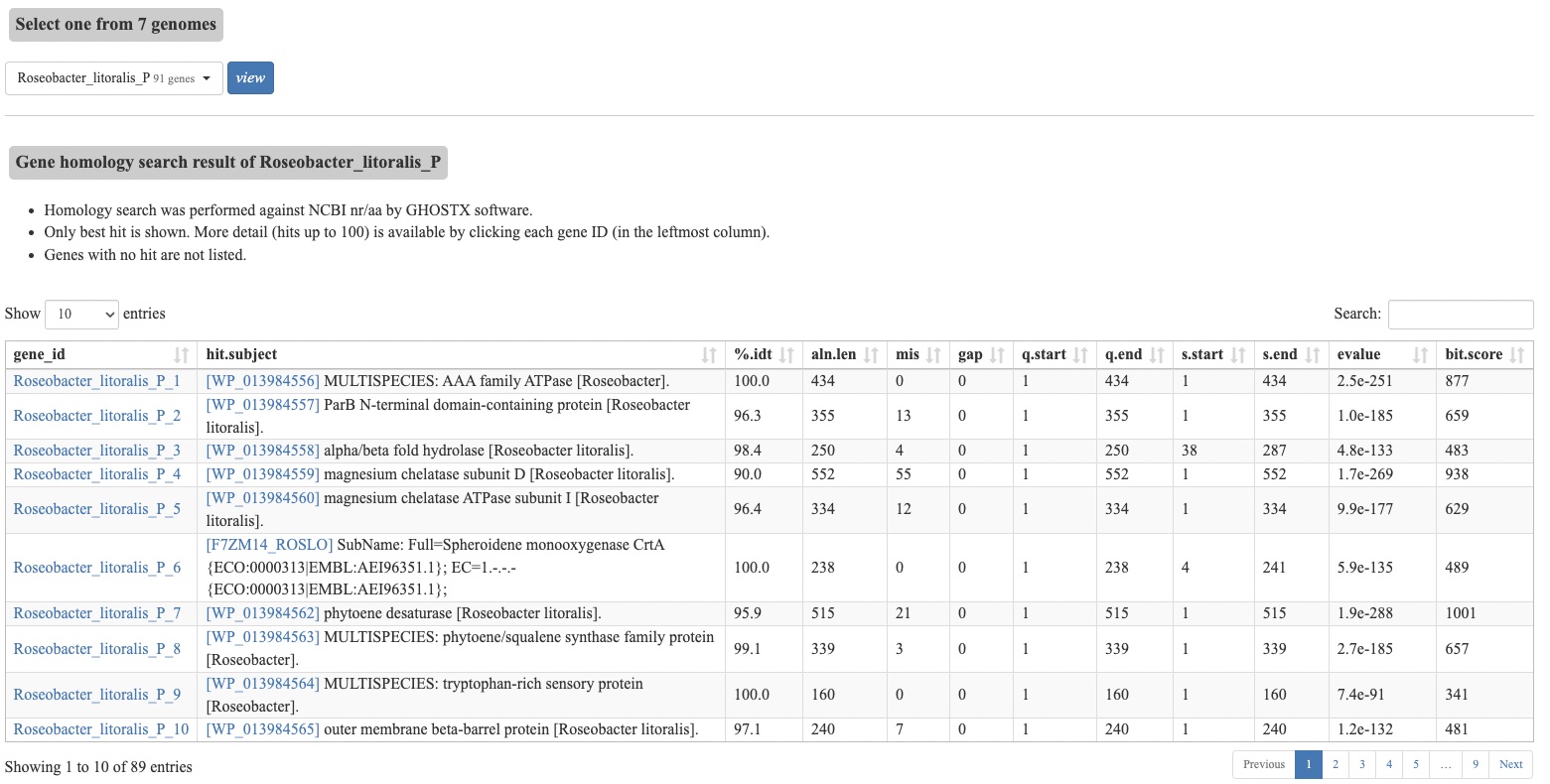
Browsing gene annotation
About
DiGAlign is a genome alignment viewer that extends the alignment visualizing function implemented in ViPTree. Nucleotide- (BLASTn) or amino acid- (tBLASTx) resolved alignments of input nucleotide sequences are computed and interactively visualized to generate a publication-ready figure. Same as ViPTree, the alignment view can include gene and function predictions performed by the DiGAlign server or provided by the user. DiGAlign quickly and effectively assists you in visualizing the results as you want in comparative genomics and genome structure exploration.
Workflow
Example
Here is an exmaple of the session main page (an entrance page after the computation).
Snapshots
Version history
Citation
If you use results (data / figures) generated by DiGAlign in your research, please cite:Feedback
We appreciate bug reports, comments, and suggestions. The form is here.