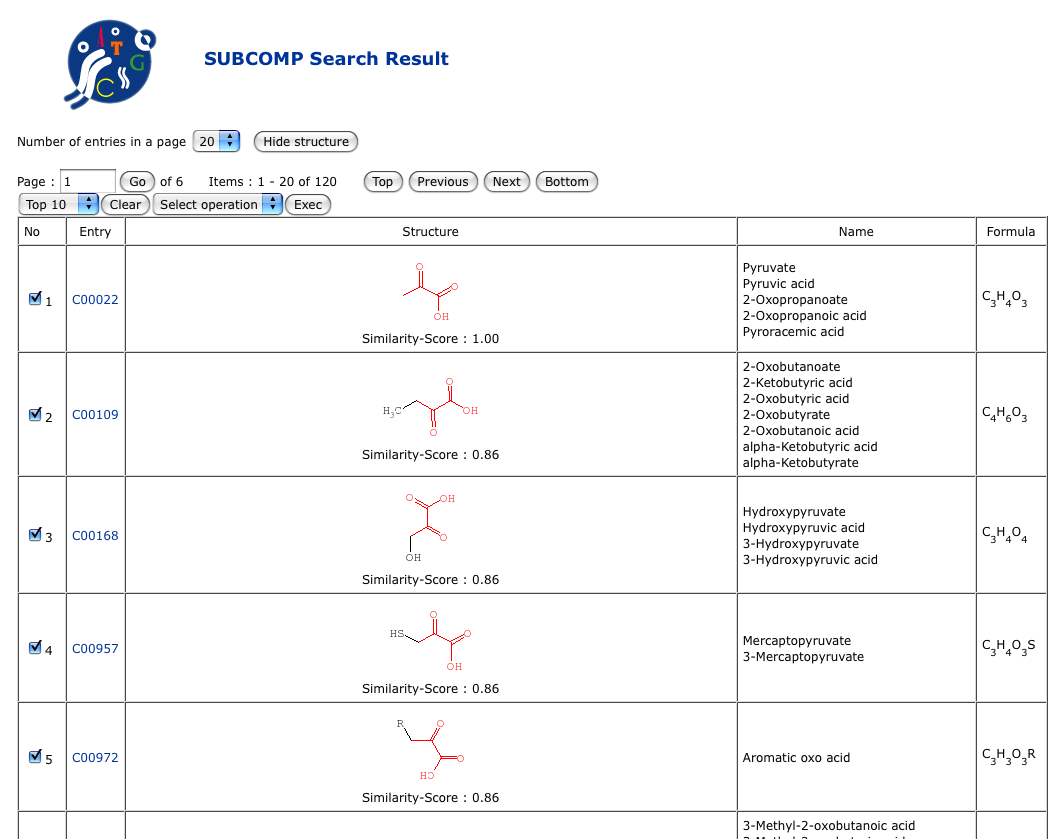
About SUBCOMPSUBCOMP (SUBstructure matching of COMPounds) is a bit-string based method for comparing chemical structures. It has been implemented in the KEGG system for searching similar chemical structures in the chemical structure databases, often as a rapid alternative to more time-consuming (but more accurate) SIMCOMP.SUBCOMP identifies database compounds containing substructures that match the entire query compound (superstructures of query compound) and database compounds that match any substructure of the query compound (substructures of query compound).
SUBCOMP provides the atom alignments between two chemical compound graphs, then it can also calculate the similarity of two chemical compounds based on the same equation as that of SIMCOMP. All computations of SUBCOMP are done with the MDL/Mol file format. The current version of GenomeNet structural search provides the availabilities for not only MDL/Mol file format but also SMILES format. When SMILES format is input, it will be transformed into MDL/Mol formats internally. How to use the query formHow to use the advanced optionsThe current version of SUBCOMP offers another search options named as the "Advanced options", which will be shown when clicking the small triangle ( ). ).
"Search mode" is which type of substructures should be found. To check "SUBstructure" or "SUPERstructure" is to find database compounds which are containing the entire query compound or are entirely contained within the query compound. "All" means to find both types of substructures simultaneously. Several conditions to match atoms are available as "Matching conditions": - Check "Charge" to distinguish ionized atoms from normal atoms. For instance, sometimes the oxygen of a hydroxyl group ("OH") is described as the anion like "O-". - Check "Valence" to distinguish the valence of each atom, in other words, the oxidation state of each atom. - Check "Coordinate bond" to consider the coordinate bond formed between anion-cation single bond. In this case, such a bond is just treated as a normal double bond. - Check "Chiral" to distinguish any isomers, provided that the information of chirality is properly given in Mol format. Here the R-/S- chirality of asymmetric carbons can be designated as the up- or down- arrows on the 2D graph, and the cis-/trans- chirality around C=C double bond should be described with proper coordinates on 2D plane. Further operation
Reference - Hattori, M., Tanaka, N., Kanehisa, M., and Goto, S.; SIMCOMP/SUBCOMP: chemical structure search servers for network analyses. Nucleic Acids Res. 38, W652-W656 (2010). [pubmed] Last updated: August 20, 2010 « Back |