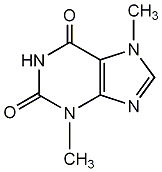
Metabolite
KNApSAcK Entry
| id | C00001509 |
|---|---|
| Name | Theobromine |
| CAS RN | 83-67-0 |
| Standard InChI | InChI=1S/C7H8N4O2/c1-10-3-8-5-4(10)6(12)9-7(13)11(5)2/h3H,1-2H3,(H,9,12,13) |
| Standard InChI (Main Layer) | InChI=1S/C7H8N4O2/c1-10-3-8-5-4(10)6(12)9-7(13)11(5)2/h3H,1-2H3,(H,9,12,13) |
Cluster
| Phytochemical cluster | No. 12 |
|---|---|
| KCF-S cluster | No. 2266 |
Link
ChEMBL
| By standard InChI | CHEMBL1114 |
|---|---|
| By standard InChI Main Layer | CHEMBL1114 |
KEGG
| By LinkDB | C07480 |
|---|
CTD
| By CAS RN | D013805 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 10 |
| asterids | 7 |
| eudicotyledons | 1 |
Family
| family name | count |
|---|---|
| Theaceae | 5 |
| Sapindaceae | 4 |
| Rutaceae | 3 |
| Aquifoliaceae | 2 |
| Malvaceae | 2 |
| Fabaceae | 1 |
| Loranthaceae | 1 |
List (18)
* NCBI
Human Protein / Gene in interaction
13 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | CHEMBL1114 |
CHEMBL1741321
(1)
|
1 / 0 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | CHEMBL1114 |
CHEMBL1794573
(1)
|
2 / 2 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | CHEMBL1114 |
CHEMBL1738600
(1)
|
0 / 0 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | CHEMBL1114 |
CHEMBL1794371
(1)
|
0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | CHEMBL1114 |
CHEMBL1741325
(1)
|
0 / 1 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | CHEMBL1114 |
CHEMBL1738610
(1)
|
0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | CHEMBL1114 |
CHEMBL1794467
(1)
|
0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | CHEMBL1114 |
CHEMBL1613808
(1)
|
0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | CHEMBL1114 |
CHEMBL1741322
(1)
|
0 / 0 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | CHEMBL1114 |
CHEMBL1614171
(1)
|
5 / 2 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | CHEMBL1114 |
CHEMBL1741323
(1)
|
1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL1114 |
CHEMBL1614108
(1)
CHEMBL1613886
(1)
CHEMBL1741324 (1) |
0 / 1 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | CHEMBL1114 |
CHEMBL1738442
(2)
|
0 / 0 |
CTD interaction (7)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D013805 | 1544 |
CYP1A2
CP12 P3-450 P450(PA) |
cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) | CYP1A2 protein results in increased chemical synthesis of Theobromine |
increases chemical synthesis
|
protein |
1302044
|
| D013805 | 1544 |
CYP1A2
CP12 P3-450 P450(PA) |
cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) | CYP1A2 protein results in increased metabolism of Theobromine |
increases metabolic processing
|
protein |
10215755
|
| D013805 | 1544 |
CYP1A2
CP12 P3-450 P450(PA) |
cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) | furafylline affects the reaction [CYP1A2 protein results in increased metabolism of Theobromine] |
affects reaction
/ increases metabolic processing |
protein |
10215755
|
| D013805 | 1571 |
CYP2E1
CPE1 CYP2E P450-J P450C2E |
cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) | 4-nitrophenol inhibits the reaction [CYP2E1 protein results in increased metabolism of Theobromine] |
decreases reaction
/ increases metabolic processing |
protein |
10215755
|
| D013805 | 1571 |
CYP2E1
CPE1 CYP2E P450-J P450C2E |
cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) | CYP2E1 protein results in increased chemical synthesis of Theobromine |
increases chemical synthesis
|
protein |
1302044
|
| D013805 | 1571 |
CYP2E1
CPE1 CYP2E P450-J P450C2E |
cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) | CYP2E1 protein results in increased metabolism of Theobromine |
increases metabolic processing
|
protein |
10215755
|
| D013805 | 1571 |
CYP2E1
CPE1 CYP2E P450-J P450C2E |
cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) | Ditiocarb inhibits the reaction [CYP2E1 protein results in increased metabolism of Theobromine] |
decreases reaction
/ increases metabolic processing |
protein |
10215755
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (9)
| OMIM | preferred title | UniProt |
|---|---|---|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
KEGG DISEASE (7)
| KEGG | disease name | UniProt |
|---|---|---|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
Q01453
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|