About KAAS
KAAS (KEGG Automatic Annotation Server) provides functional annotation of genes by BLAST or GHOST comparisons against the manually curated KEGG GENES database. The result contains KO (KEGG Orthology) assignments and automatically generated KEGG pathways.
Complete or Draft Genome
KAAS works best when a complete set of genes in a genome is known. Prepare query amino acid sequences and use the BBH (bi-directional best hit) method to assign orthologs.
Partial Genome
KAAS can also be used for a limited number of genes. Prepare query amino acid sequences and use the SBH (single-directional best hit) method to assign orthologs.
Metagenomes
When the query consisits of large numbers of sequences and / or sequences from mixture of species such as those from metagenome sequencing project, we recommend the GHOSTX search and SBH method.
Example of Results
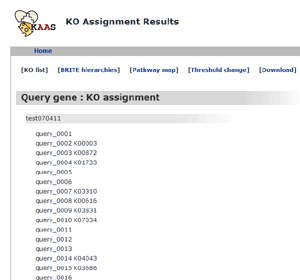
KO assignment
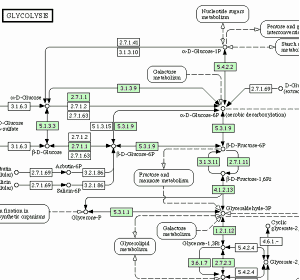
KEGG pathway mapping