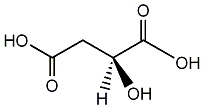
Metabolite
KNApSAcK Entry
| id | C00001192 |
|---|---|
| Name | L-Malic acid / L-(-)-Malic acid |
| CAS RN | 97-67-6 |
| Standard InChI | InChI=1S/C4H6O5/c5-2(4(8)9)1-3(6)7/h2,5H,1H2,(H,6,7)(H,8,9)/t2-/m0/s1 |
| Standard InChI (Main Layer) | InChI=1S/C4H6O5/c5-2(4(8)9)1-3(6)7/h2,5H,1H2,(H,6,7)(H,8,9) |
Cluster
| Phytochemical cluster | |
|---|---|
| KCF-S cluster | No. 8373 |
Link
ChEMBL
| By standard InChI | |
|---|---|
| By standard InChI Main Layer | CHEMBL225986 CHEMBL1455497 |
KEGG
| By LinkDB | C00149 |
|---|
CTD
| By CAS RN |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 1 |
| eudicotyledons | 1 |
Family
| family name | count |
|---|---|
| Brassicaceae | 1 |
| Enterobacteriaceae | 1 |
| Crassulaceae | 1 |
List (4)
* NCBI
| KNApSAcK organism | *ID | *family | *plant class | *kingdom |
|---|---|---|---|---|
| Arabidopsis thaliana | 3702 | Brassicaceae | rosids | Viridiplantae |
| Escherichia coli | 562 | Enterobacteriaceae | Bacteria | |
| Kalanchoe daigremontiana | 23013 | Crassulaceae | eudicotyledons | Viridiplantae |
| Opunitia dillenii HAW. |
Human Protein / Gene in interaction
12 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | CHEMBL1455497 |
CHEMBL1741321
(1)
|
1 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | CHEMBL225986 |
CHEMBL1613842
(1)
|
4 / 2 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | CHEMBL1455497 |
CHEMBL1738312
(1)
|
0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | CHEMBL1455497 |
CHEMBL1741325
(1)
|
0 / 1 |
| Q9GZT9 | Egl nine homolog 1 | Enzyme | CHEMBL1455497 |
CHEMBL2318597
(1)
|
1 / 1 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | CHEMBL1455497 |
CHEMBL1741322
(1)
|
0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | CHEMBL1455497 |
CHEMBL1741323
(1)
|
1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL1455497 |
CHEMBL1741324
(1)
|
0 / 1 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | CHEMBL1455497 |
CHEMBL1737991
(1)
|
0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | CHEMBL1455497 |
CHEMBL1738442
(1)
|
0 / 0 |
| Q9C0B1 | Alpha-ketoglutarate-dependent dioxygenase FTO | Unclassified protein | CHEMBL1455497 |
CHEMBL2344107
(1)
|
1 / 2 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | CHEMBL225986 |
CHEMBL2354287
(1)
|
1 / 1 |
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (9)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #609820 | Erythrocytosis, familial, 3; ecyt3 |
Q9GZT9
|
| #612938 | Growth retardation, developmental delay, coarse facies, and early death; gdfd |
Q9C0B1
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
KEGG DISEASE (8)
| KEGG | disease name | UniProt |
|---|---|---|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
Q13148
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
Q16637 (related) |
| H00409 | Type II diabetes mellitus |
Q9C0B1
(related)
|
| H00926 | Growth retardation, developmental delay, coarse facies, and early death |
Q9C0B1
(related)
|
| H00236 | Congenital polycythemia |
Q9GZT9
(related)
|