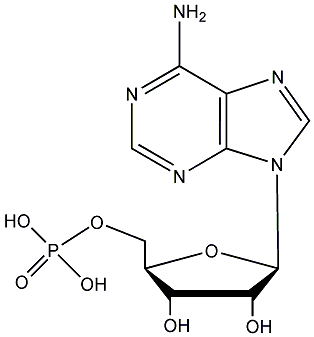
Metabolite
KNApSAcK Entry
| id | C00019347 |
|---|---|
| Name | AMP / Adenosine 5'-phosphate |
| CAS RN | 61-19-8 |
| Standard InChI | InChI=1S/C10H14N5O7P/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(22-10)1-21-23(18,19)20/h2-4,6-7,10,16-17H,1H2,(H2,11,12,13)(H2,18,19,20)/t4-,6?,7+,10-/m1/s1 |
| Standard InChI (Main Layer) | InChI=1S/C10H14N5O7P/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(22-10)1-21-23(18,19)20/h2-4,6-7,10,16-17H,1H2,(H2,11,12,13)(H2,18,19,20) |
Cluster
| Phytochemical cluster | |
|---|---|
| KCF-S cluster | No. 1201 |
Link
ChEMBL
| By standard InChI | |
|---|---|
| By standard InChI Main Layer | CHEMBL16720 CHEMBL752 CHEMBL603811 CHEMBL608290 CHEMBL1230732 |
KEGG
| By LinkDB | C00020 |
|---|
CTD
| By CAS RN | D000249 |
|---|
Species
Summary
Plant class
| class name | count |
|---|
Family
| family name | count |
|---|---|
| Enterobacteriaceae | 1 |
List (1)
* NCBI
| KNApSAcK organism | *ID | *family | *plant class | *kingdom |
|---|---|---|---|---|
| Escherichia coli | 562 | Enterobacteriaceae | Bacteria |
Human Protein / Gene in interaction
21 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| Q16637 | Survival motor neuron protein | Unclassified protein | CHEMBL16720 |
CHEMBL1613842
(1)
|
4 / 2 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | CHEMBL752 |
CHEMBL1794499
(1)
|
2 / 0 |
| P47900 | P2Y purinoceptor 1 | Purine receptor | CHEMBL752 |
CHEMBL750157
(2)
|
0 / 0 |
| P21589 | 5'-nucleotidase | Enzyme | CHEMBL752 |
CHEMBL1042900
(1)
|
1 / 1 |
| P51582 | P2Y purinoceptor 4 | Purine receptor | CHEMBL752 |
CHEMBL756884
(1)
CHEMBL757041
(1)
|
0 / 0 |
| P60891 | Ribose-phosphate pyrophosphokinase 1 | Enzyme | CHEMBL603811 |
CHEMBL760863
(1)
CHEMBL760865
(1)
CHEMBL760868 (1) |
4 / 4 |
| Q96G91 | P2Y purinoceptor 11 | Purine receptor | CHEMBL752 |
CHEMBL755046
(1)
CHEMBL756287
(1)
|
0 / 0 |
| P41231 | P2Y purinoceptor 2 | Purine receptor | CHEMBL752 |
CHEMBL753285
(2)
|
0 / 0 |
| P07195 | L-lactate dehydrogenase B chain | Enzyme | CHEMBL752 |
CHEMBL2060672
(1)
|
1 / 4 |
| P05186 | Alkaline phosphatase, tissue-nonspecific isozyme | Enzyme | CHEMBL752 |
CHEMBL1042901
(1)
|
3 / 1 |
| P00338 | L-lactate dehydrogenase A chain | Enzyme | CHEMBL752 |
CHEMBL2060668
(1)
CHEMBL2060670
(1)
CHEMBL2060671 (1) |
1 / 5 |
| P12931 | Proto-oncogene tyrosine-protein kinase Src | Src | CHEMBL752 |
CHEMBL942401
(1)
|
0 / 0 |
| P09467 | Fructose-1,6-bisphosphatase 1 | Enzyme | CHEMBL752 |
CHEMBL694959
(1)
CHEMBL981259
(1)
CHEMBL1074684 (1) CHEMBL1074685 (1) CHEMBL1106396 (1) |
1 / 1 |
| Q6DHV7 | Adenosine deaminase-like protein | Enzyme | CHEMBL752 |
CHEMBL1817385
(1)
CHEMBL1817386
(2)
|
0 / 0 |
| P30542 | Adenosine receptor A1 | Adenosine receptor | CHEMBL752 |
CHEMBL2169616
(1)
|
0 / 0 |
| P28329 | Choline O-acetyltransferase | Enzyme | CHEMBL752 |
CHEMBL662975
(1)
|
1 / 1 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | CHEMBL752 |
CHEMBL1738442
(1)
|
0 / 0 |
| Q9UBF8 | Phosphatidylinositol 4-kinase beta | Enzyme | CHEMBL752 |
CHEMBL758629
(1)
|
0 / 0 |
| Q8TCG2 | Phosphatidylinositol 4-kinase type 2-beta | Enzyme | CHEMBL752 |
CHEMBL758629
(1)
|
0 / 0 |
| Q9BTU6 | Phosphatidylinositol 4-kinase type 2-alpha | Enzyme | CHEMBL752 |
CHEMBL758629
(1)
|
0 / 0 |
| P42356 | Phosphatidylinositol 4-kinase alpha | Enzyme | CHEMBL752 |
CHEMBL758629
(1)
|
0 / 0 |
CTD interaction (6)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D000249 | 216 |
ALDH1A1
ALDC ALDH-E1 ALDH1 ALDH11 PUMB1 RALDH1 |
aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) | [4-dimethylaminocinnamaldehyde co-treated with Adenosine Monophosphate] binds to ALDH1A1 protein |
affects binding
/ affects cotreatment |
protein |
10609638
|
| D000249 | 170685 |
NUDT10
APS2 DIPP3a hDIPP3alpha |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 (EC:3.6.1.52 3.6.1.60) | [NUDT10 protein results in increased metabolism of diadenosine 5',5''''-P1,P6-hexaphosphate] which results in increased chemical synthesis of Adenosine Monophosphate |
increases chemical synthesis
/ increases metabolic processing |
protein |
12121577
|
| D000249 | 170685 |
NUDT10
APS2 DIPP3a hDIPP3alpha |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 (EC:3.6.1.52 3.6.1.60) | [NUDT10 protein results in increased metabolism of P(1),P(5)-di(adenosine-5'-)pentaphosphate] which results in increased chemical synthesis of Adenosine Monophosphate |
increases chemical synthesis
/ increases metabolic processing |
protein |
12121577
|
| D000249 | 55190 |
NUDT11
APS1 ASP1 DIPP3b DIPP3beta hDIPP3beta |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 (EC:3.6.1.52 3.6.1.60) | [NUDT11 protein results in increased metabolism of diadenosine 5',5''''-P1,P6-hexaphosphate] which results in increased chemical synthesis of Adenosine Monophosphate |
increases chemical synthesis
/ increases metabolic processing |
protein |
12121577
|
| D000249 | 55190 |
NUDT11
APS1 ASP1 DIPP3b DIPP3beta hDIPP3beta |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 (EC:3.6.1.52 3.6.1.60) | [NUDT11 protein results in increased metabolism of P(1),P(5)-di(adenosine-5'-)pentaphosphate] which results in increased chemical synthesis of Adenosine Monophosphate |
increases chemical synthesis
/ increases metabolic processing |
protein |
12121577
|
| D000249 | 10891 |
PPARGC1A
LEM6 PGC-1(alpha) PGC-1v PGC1 PGC1A PPARGC1 |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | Adenosine Monophosphate results in increased expression of PPARGC1A mRNA |
increases expression
|
mRNA |
23056435
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (18)
| OMIM | preferred title | UniProt |
|---|---|---|
| #301835 | Arts syndrome; arts |
P60891
|
| #211800 | Calcification of joints and arteries; calja |
P21589
|
| #311070 | Charcot-marie-tooth disease, x-linked recessive, 5; cmtx5 |
P60891
|
| #304500 | Deafness, x-linked 1; dfnx1 |
P60891
|
| #229700 | Fructose-1,6-bisphosphatase deficiency |
P09467
|
| #612933 | Glycogen storage disease xi; gsd11 |
P00338
|
| #146300 | Hypophosphatasia, adult |
P05186
|
| #241510 | Hypophosphatasia, childhood |
P05186
|
| #241500 | Hypophosphatasia, infantile |
P05186
|
| #614128 | Lactate dehydrogenase b deficiency; ldhbd |
P07195
|
| #254210 | Myasthenic syndrome, congenital, associated with episodic apnea |
P28329
|
| #300661 | Phosphoribosylpyrophosphate synthetase superactivity |
P60891
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
KEGG DISEASE (14)
| KEGG | disease name | UniProt |
|---|---|---|
| H00069 | Glycogen storage diseases (GSD) |
P00338
(related)
|
| H00010 | Multiple myeloma |
P00338
(marker)
P07195 (marker) |
| H00023 | Testicular cancer |
P00338
(marker)
P07195 (marker) |
| H00035 | Ewing's sarcoma |
P00338
(marker)
P07195 (marker) |
| H00043 | Neuroblastoma |
P00338
(marker)
P07195 (marker) |
| H00213 | Hypophosphatasia |
P05186
(related)
|
| H00114 | Fructose-1,6-bisphosphatase deficiency |
P09467
(related)
|
| H00824 | Calcification of joints and arteries |
P21589
(related)
|
| H00770 | Congenital myasthenic syndrome |
P28329
(related)
|
| H00196 | Phosphoribosylpyrophosphate synthetase I superactivity |
P60891
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P60891
(related)
|
| H00946 | Arts syndrome |
P60891
(related)
|
| H01209 | Deafness, X-linked |
P60891
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
Q16637 (related) |