Metabolite
KNApSAcK Entry
| id | C00007230 |
|---|---|
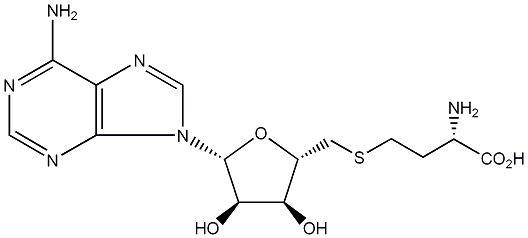
| Name | S-Adenosyl-L-homocysteine |
| CAS RN | 979-92-0 |
| Standard InChI | InChI=1S/C14H20N6O5S/c15-6(14(23)24)1-2-26-3-7-9(21)10(22)13(25-7)20-5-19-8-11(16)17-4-18-12(8)20/h4-7,9-10,13,21-22H,1-3,15H2,(H,23,24)(H2,16,17,18)/t6-,7+,9+,10+,13+/m0/s1 |
| Standard InChI (Main Layer) | InChI=1S/C14H20N6O5S/c15-6(14(23)24)1-2-26-3-7-9(21)10(22)13(25-7)20-5-19-8-11(16)17-4-18-12(8)20/h4-7,9-10,13,21-22H,1-3,15H2,(H,23,24)(H2,16,17,18) |
Cluster
| Phytochemical cluster | |
|---|---|
| KCF-S cluster | No. 5509 |
Link
ChEMBL
| By standard InChI | CHEMBL418052 |
|---|---|
| By standard InChI Main Layer | CHEMBL418052 CHEMBL45041 CHEMBL142828 CHEMBL597532 CHEMBL2092997 |
KEGG
| By LinkDB | C00021 |
|---|
CTD
| By CAS RN | D012435 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 1 |
Family
| family name | count |
|---|---|
| Brassicaceae | 1 |
| Enterobacteriaceae | 1 |
List (2)
* NCBI
| KNApSAcK organism | *ID | *family | *plant class | *kingdom |
|---|---|---|---|---|
| Arabidopsis thaliana | 3702 | Brassicaceae | rosids | Viridiplantae |
| Escherichia coli | 562 | Enterobacteriaceae | Bacteria |
Human Protein / Gene in interaction
10 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| Q86X55 | Histone-arginine methyltransferase CARM1 | Enzyme | CHEMBL418052 |
CHEMBL2175613
(1)
|
0 / 0 |
| Q8TEK3 | Histone-lysine N-methyltransferase, H3 lysine-79 specific | Enzyme | CHEMBL418052 |
CHEMBL2175606
(1)
CHEMBL2175607
(1)
CHEMBL2175616 (1) CHEMBL2350637 (1) CHEMBL2350651 (1) |
0 / 0 |
| P49327 | Fatty acid synthase | Transferase | CHEMBL418052 CHEMBL45041 |
CHEMBL682484
(2)
|
0 / 0 |
| O43463 | Histone-lysine N-methyltransferase SUV39H1 | Enzyme | CHEMBL418052 |
CHEMBL2175614
(1)
|
0 / 0 |
| Q9UBC3 | DNA (cytosine-5)-methyltransferase 3B | Enzyme | CHEMBL418052 |
CHEMBL1036409
(1)
CHEMBL1036445
(1)
CHEMBL1072865 (1) CHEMBL1072867 (1) |
1 / 1 |
| P50135 | Histamine N-methyltransferase | Enzyme | CHEMBL418052 CHEMBL2092997 |
CHEMBL696454
(2)
|
0 / 0 |
| Q99873 | Protein arginine N-methyltransferase 1 | Enzyme | CHEMBL418052 |
CHEMBL2175615
(1)
|
0 / 0 |
| P26358 | DNA (cytosine-5)-methyltransferase 1 | Transferase | CHEMBL418052 |
CHEMBL1036408
(1)
CHEMBL1036444
(1)
CHEMBL1072864 (1) CHEMBL1072866 (1) CHEMBL1918222 (1) CHEMBL1918223 (1) CHEMBL2019574 (1) CHEMBL2050426 (1) CHEMBL2050428 (1) |
2 / 0 |
| O95050 | Indolethylamine N-methyltransferase | Enzyme | CHEMBL418052 |
CHEMBL696703
(1)
|
0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | CHEMBL597532 |
CHEMBL1614421
(1)
|
4 / 3 |
CTD interaction (4)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D012435 | 1277 |
COL1A1
OI4 |
collagen, type I, alpha 1 | S-Adenosylhomocysteine results in decreased expression of COL1A1 mRNA |
decreases expression
|
mRNA |
15983038
|
| D012435 | 1278 |
COL1A2
OI4 |
collagen, type I, alpha 2 | S-Adenosylhomocysteine results in decreased expression of COL1A2 mRNA |
decreases expression
|
mRNA |
15983038
|
| D012435 | 1786 |
DNMT1
ADCADN AIM CXXC9 DNMT HSN1E MCMT |
DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) | S-Adenosylhomocysteine results in decreased activity of DNMT1 protein |
decreases activity
|
protein |
16037419
|
| D012435 | 10891 |
PPARGC1A
LEM6 PGC-1(alpha) PGC-1v PGC1 PGC1A PPARGC1 |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | S-Adenosylhomocysteine results in increased expression of PPARGC1A mRNA |
increases expression
|
mRNA |
23056435
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (7)
| OMIM | preferred title | UniProt |
|---|---|---|
| #604121 | Cerebellar ataxia, deafness, and narcolepsy, autosomal dominant; adcadn |
P26358
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #242860 | Immunodeficiency-centromeric instability-facial anomalies syndrome 1; icf1 |
Q9UBC3
|
| #614116 | Neuropathy, hereditary sensory, type ie; hsn1e |
P26358
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|