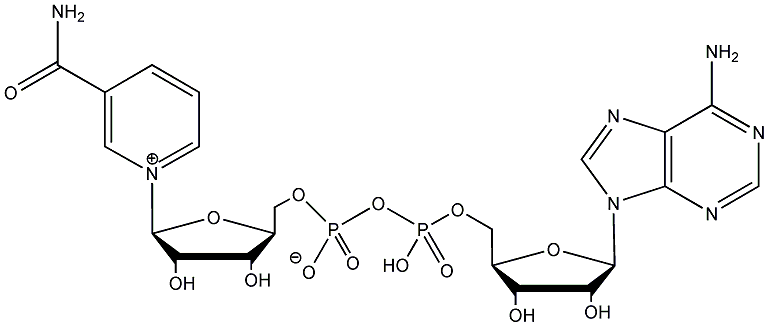
Metabolite
KNApSAcK Entry
| id | C00007256 |
|---|---|
| Name | NAD / Nicotinamide adenine dinucleotide |
| CAS RN | 53-84-9 |
| Standard InChI | InChI=1S/C21H27N7O14P2/c22-17-12-19(25-7-24-17)28(8-26-12)21-16(32)14(30)11(41-21)6-39-44(36,37)42-43(34,35)38-5-10-13(29)15(31)20(40-10)27-3-1-2-9(4-27)18(23)33/h1-4,7-8,10-11,13-16,20-21,29-32H,5-6H2,(H5-,22,23,24,25,33,34,35,36,37)/t10-,11+,13+,14?,15?,16-,20-,21+/m0/s1 |
| Standard InChI (Main Layer) | InChI=1S/C21H27N7O14P2/c22-17-12-19(25-7-24-17)28(8-26-12)21-16(32)14(30)11(41-21)6-39-44(36,37)42-43(34,35)38-5-10-13(29)15(31)20(40-10)27-3-1-2-9(4-27)18(23)33/h1-4,7-8,10-11,13-16,20-21,29-32H,5-6H2,(H5-,22,23,24,25,33,34,35,36,37) |
Cluster
| Phytochemical cluster | No. 1 |
|---|---|
| KCF-S cluster | No. 1656 |
Link
ChEMBL
| By standard InChI | |
|---|---|
| By standard InChI Main Layer | CHEMBL234870 CHEMBL1234613 CHEMBL1402028 CHEMBL1454168 CHEMBL1517480 |
KEGG
| By LinkDB | C00003 |
|---|
CTD
| By CAS RN | D009243 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 1 |
Family
| family name | count |
|---|---|
| Brassicaceae | 1 |
| Enterobacteriaceae | 1 |
List (2)
* NCBI
| KNApSAcK organism | *ID | *family | *plant class | *kingdom |
|---|---|---|---|---|
| Arabidopsis thaliana | 3702 | Brassicaceae | rosids | Viridiplantae |
| Escherichia coli | 562 | Enterobacteriaceae | Bacteria |
Human Protein / Gene in interaction
18 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | CHEMBL1402028 |
CHEMBL1738312
(1)
|
0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | CHEMBL1402028 |
CHEMBL1794499
(1)
|
2 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | CHEMBL1402028 |
CHEMBL1794585
(1)
|
0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | CHEMBL1517480 |
CHEMBL1614544
(1)
|
11 / 10 |
| P37840 | Alpha-synuclein | Unclassified protein | CHEMBL1402028 |
CHEMBL2354282
(1)
|
4 / 2 |
| P12268 | Inosine-5'-monophosphate dehydrogenase 2 | Oxidoreductase | CHEMBL1234613 |
CHEMBL699569
(1)
CHEMBL699570
(1)
|
0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | CHEMBL1402028 |
CHEMBL1794311
(1)
|
2 / 3 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | CHEMBL1402028 |
CHEMBL1794495
(1)
|
2 / 2 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | CHEMBL1517480 |
CHEMBL1738606
(1)
|
0 / 0 |
| P00338 | L-lactate dehydrogenase A chain | Enzyme | CHEMBL1234613 |
CHEMBL859428
(1)
|
1 / 5 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | CHEMBL1402028 |
CHEMBL1738317
(1)
|
0 / 0 |
| P34981 | Thyrotropin-releasing hormone receptor | Neuropeptide receptor | CHEMBL1402028 |
CHEMBL1737990
(1)
|
0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | CHEMBL1402028 |
CHEMBL1737991
(1)
|
0 / 0 |
| P07327 | Alcohol dehydrogenase 1A | Oxidoreductase | CHEMBL1234613 |
CHEMBL645400
(1)
|
0 / 0 |
| P28329 | Choline O-acetyltransferase | Enzyme | CHEMBL1234613 |
CHEMBL662977
(1)
|
1 / 1 |
| O95544 | NAD kinase | Enzyme | CHEMBL234870 |
CHEMBL891091
(1)
CHEMBL891095
(1)
|
0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | CHEMBL1402028 |
CHEMBL2354311
(1)
|
1 / 0 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | CHEMBL1402028 |
CHEMBL2354287
(1)
|
1 / 1 |
CTD interaction (48)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D009243 | 10327 |
AKR1A1
ALDR1 ALR ARM DD3 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) (EC:1.1.1.2) | NAD promotes the reaction [AKR1A1 protein affects the chemical synthesis of benzo(a)pyrene-7,8-dione] |
affects chemical synthesis
/ increases reaction |
protein |
15720144
|
| D009243 | 10327 |
AKR1A1
ALDR1 ALR ARM DD3 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) (EC:1.1.1.2) | NAD promotes the reaction [AKR1A1 protein affects the metabolism of and results in increased activity of benzo(a)pyrene 7,8-dihydrodiol] |
affects metabolic processing
/ increases activity / increases reaction |
protein |
15720144
|
| D009243 | 216 |
ALDH1A1
ALDC ALDH-E1 ALDH1 ALDH11 PUMB1 RALDH1 |
aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) | [ALDH1A1 protein co-treated with NAD] results in increased oxidation of aldophosphamide |
affects cotreatment
/ increases oxidation |
protein |
10856427
|
| D009243 | 216 |
ALDH1A1
ALDC ALDH-E1 ALDH1 ALDH11 PUMB1 RALDH1 |
aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) | [ALDH1A1 protein co-treated with NAD] results in increased oxidation of propionaldehyde |
affects cotreatment
/ increases oxidation |
protein |
10559215
10856427 |
| D009243 | 216 |
ALDH1A1
ALDC ALDH-E1 ALDH1 ALDH11 PUMB1 RALDH1 |
aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) | [ALDH1A1 protein co-treated with NAD] results in increased oxidation of Retinaldehyde |
affects cotreatment
/ increases oxidation |
protein |
10559215
|
| D009243 | 216 |
ALDH1A1
ALDC ALDH-E1 ALDH1 ALDH11 PUMB1 RALDH1 |
aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) | [NAD co-treated with propionaldehyde] inhibits the reaction [Acrolein results in decreased activity of ALDH1A1 protein] |
affects cotreatment
/ decreases activity / decreases reaction |
protein |
22339434
|
| D009243 | 216 |
ALDH1A1
ALDC ALDH-E1 ALDH1 ALDH11 PUMB1 RALDH1 |
aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) | NAD inhibits the reaction [Acrolein binds to and results in decreased activity of ALDH1A1 protein] |
affects binding
/ decreases activity / decreases reaction |
protein |
22339434
|
| D009243 | 219 |
ALDH1B1
ALDH5 ALDHX |
aldehyde dehydrogenase 1 family, member B1 (EC:1.2.1.3) | [ALDH1B1 protein co-treated with NAD] results in increased metabolism of Acetaldehyde |
affects cotreatment
/ increases metabolic processing |
protein |
7779080
|
| D009243 | 219 |
ALDH1B1
ALDH5 ALDHX |
aldehyde dehydrogenase 1 family, member B1 (EC:1.2.1.3) | [ALDH1B1 protein co-treated with NAD] results in increased metabolism of propionaldehyde |
affects cotreatment
/ increases metabolic processing |
protein |
7779080
|
| D009243 | 217 |
ALDH2
ALDH-E2 ALDHI ALDM |
aldehyde dehydrogenase 2 family (mitochondrial) (EC:1.2.1.3) | [ALDH2 protein co-treated with NAD] results in increased oxidation of Retinaldehyde |
affects cotreatment
/ increases oxidation |
protein |
10559215
|
| D009243 | 217 |
ALDH2
ALDH-E2 ALDHI ALDM |
aldehyde dehydrogenase 2 family (mitochondrial) (EC:1.2.1.3) | NAD inhibits the reaction [Acrolein binds to and results in decreased activity of ALDH2 protein] |
affects binding
/ decreases activity / decreases reaction |
protein |
22339434
|
| D009243 | 218 |
ALDH3A1
ALDH3 ALDHIII |
aldehyde dehydrogenase 3 family, member A1 (EC:1.2.1.5) | [ALDH3A1 protein co-treated with NAD] results in increased oxidation of aldophosphamide |
affects cotreatment
/ increases oxidation |
protein |
10856427
|
| D009243 | 218 |
ALDH3A1
ALDH3 ALDHIII |
aldehyde dehydrogenase 3 family, member A1 (EC:1.2.1.5) | [ALDH3A1 protein co-treated with NAD] results in increased oxidation of benzaldehyde |
affects cotreatment
/ increases oxidation |
protein |
10856427
|
| D009243 | 581 |
BAX
BCL2L4 |
BCL2-associated X protein | NAD results in decreased expression of BAX protein |
decreases expression
|
protein |
12390773
|
| D009243 | 596 |
BCL2
Bcl-2 PPP1R50 |
B-cell CLL/lymphoma 2 | NAD results in increased expression of BCL2 protein |
increases expression
|
protein |
12390773
|
| D009243 | 847 |
CAT
|
catalase (EC:1.11.1.6) | CAT protein inhibits the reaction [[2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of HRAS1 gene] |
affects cotreatment
/ decreases reaction / increases mutagenesis |
protein |
10334203
|
| D009243 | 847 |
CAT
|
catalase (EC:1.11.1.6) | CAT protein inhibits the reaction [[2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of TP53 gene] |
affects cotreatment
/ decreases reaction / increases mutagenesis |
protein |
10334203
|
| D009243 | 1050 |
CEBPA
C/EBP-alpha CEBP |
CCAAT/enhancer binding protein (C/EBP), alpha | [NAD co-treated with NQO1 protein] results in decreased degradation of CEBPA protein |
affects cotreatment
/ decreases degradation |
protein |
23086932
|
| D009243 | 1727 |
CYB5R3
B5R DIA1 |
cytochrome b5 reductase 3 (EC:1.6.2.2) | CYB5R3 protein binds to NAD |
affects binding
|
protein |
16469290
|
| D009243 | 10495 |
ENOX2
APK1 COVA1 tNOX |
ecto-NOX disulfide-thiol exchanger 2 | [epigallocatechin gallate results in decreased activity of ENOX2 protein] which results in increased abundance of NAD |
decreases activity
/ increases abundance |
protein |
20518072
|
| D009243 | 10495 |
ENOX2
APK1 COVA1 tNOX |
ecto-NOX disulfide-thiol exchanger 2 | [phenoxodiol results in decreased activity of ENOX2 protein] which results in increased abundance of NAD |
decreases activity
/ increases abundance |
protein |
20518072
|
| D009243 | 2597 |
GAPDH
G3PD GAPD |
glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12) | Adenosine Diphosphate inhibits the reaction [[GAPDH protein co-treated with NAD co-treated with Glyceraldehyde 3-Phosphate co-treated with Glutathione] results in increased reduction of sodium arsenite analog] |
affects cotreatment
/ decreases reaction / increases reduction |
protein |
15788719
|
| D009243 | 2597 |
GAPDH
G3PD GAPD |
glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12) | Adenosine Triphosphate inhibits the reaction [[GAPDH protein co-treated with NAD co-treated with Glyceraldehyde 3-Phosphate co-treated with Glutathione] results in increased reduction of sodium arsenite analog] |
affects cotreatment
/ decreases reaction / increases reduction |
protein |
15788719
|
| D009243 | 2597 |
GAPDH
G3PD GAPD |
glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12) | [GAPDH protein co-treated with NAD co-treated with Glyceraldehyde 3-Phosphate co-treated with Glutathione] results in increased reduction of sodium arsenite analog |
affects cotreatment
/ increases reduction |
protein |
15788719
|
| D009243 | 2597 |
GAPDH
G3PD GAPD |
glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12) | NAD analog inhibits the reaction [[GAPDH protein co-treated with NAD co-treated with Glyceraldehyde 3-Phosphate co-treated with Glutathione] results in increased reduction of sodium arsenite analog] |
affects cotreatment
/ decreases reaction / increases reduction |
protein |
15788719
|
| D009243 | 2597 |
GAPDH
G3PD GAPD |
glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12) | NADP inhibits the reaction [[GAPDH protein co-treated with NAD co-treated with Glyceraldehyde 3-Phosphate co-treated with Glutathione] results in increased reduction of sodium arsenite analog] |
affects cotreatment
/ decreases reaction / increases reduction |
protein |
15788719
|
| D009243 | 3033 |
HADH
HAD HADH1 HADHSC HCDH HHF4 MSCHAD SCHAD |
hydroxyacyl-CoA dehydrogenase (EC:1.1.1.35) | HADH protein results in increased metabolism of NAD |
increases metabolic processing
|
protein |
10600649
|
| D009243 | 15461 | [2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of HRAS1 gene |
affects cotreatment
/ increases mutagenesis |
gene |
10334203
|
||
| D009243 | 15461 | bathocuproine inhibits the reaction [[2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of HRAS1 gene] |
affects cotreatment
/ decreases reaction / increases mutagenesis |
gene |
10334203
|
||
| D009243 | 15461 | CAT protein inhibits the reaction [[2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of HRAS1 gene] |
affects cotreatment
/ decreases reaction / increases mutagenesis |
gene |
10334203
|
||
| D009243 | 55191 |
NADSYN1
|
NAD synthetase 1 (EC:6.3.5.1) | [NADSYN1 protein co-treated with Ammonium Chloride co-treated with nicotinic acid adenine dinucleotide co-treated with Adenosine Triphosphate] results in increased chemical synthesis of NAD |
affects cotreatment
/ increases chemical synthesis |
protein |
12547821
|
| D009243 | 1728 |
NQO1
DHQU DIA4 DTD NMOR1 NMORI QR1 |
NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) | [NAD co-treated with NQO1 protein] results in decreased degradation of CEBPA protein |
affects cotreatment
/ decreases degradation |
protein |
23086932
|
| D009243 | 142 |
PARP1
ADPRT ADPRT_1 ADPRT1 ARTD1 PARP PARP-1 PPOL pADPRT-1 |
poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) | [2,5,2',5'-tetrachlorobiphenyl results in increased activity of PARP1 protein] which results in decreased abundance of NAD |
decreases abundance
/ increases activity |
protein |
19433264
|
| D009243 | 142 |
PARP1
ADPRT ADPRT_1 ADPRT1 ARTD1 PARP PARP-1 PPOL pADPRT-1 |
poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) | [3,4,3',4'-tetrachlorobiphenyl results in increased activity of PARP1 protein] which results in decreased abundance of NAD |
decreases abundance
/ increases activity |
protein |
19433264
|
| D009243 | 142 |
PARP1
ADPRT ADPRT_1 ADPRT1 ARTD1 PARP PARP-1 PPOL pADPRT-1 |
poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) | [3-aminobenzamide results in decreased activity of PARP1 protein] inhibits the reaction [Tetrachlorodibenzodioxin results in decreased abundance of NAD] |
decreases abundance
/ decreases activity / decreases reaction |
protein |
17669606
|
| D009243 | 142 |
PARP1
ADPRT ADPRT_1 ADPRT1 ARTD1 PARP PARP-1 PPOL pADPRT-1 |
poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) | [benzamide results in decreased activity of PARP1 protein] inhibits the reaction [Tetrachlorodibenzodioxin results in decreased abundance of NAD] |
decreases abundance
/ decreases activity / decreases reaction |
protein |
17669606
|
| D009243 | 142 |
PARP1
ADPRT ADPRT_1 ADPRT1 ARTD1 PARP PARP-1 PPOL pADPRT-1 |
poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) | [coumarin results in decreased activity of PARP1 protein] inhibits the reaction [Tetrachlorodibenzodioxin results in decreased abundance of NAD] |
decreases abundance
/ decreases activity / decreases reaction |
protein |
17669606
|
| D009243 | 195814 |
SDR16C5
RDH#2 RDH-E2 RDHE2 |
short chain dehydrogenase/reductase family 16C, member 5 (EC:1.1.1.105) | NAD affects the activity of SDR16C5 protein |
affects activity
|
protein |
18926804
|
| D009243 | 23411 |
SIRT1
SIR2L1 |
sirtuin 1 | [resveratrol results in increased abundance of NAD] which results in increased expression of SIRT1 protein |
increases abundance
/ increases expression |
protein |
19928762
|
| D009243 | 6610 |
SMPD2
ISC1 NSMASE NSMASE1 |
sphingomyelin phosphodiesterase 2, neutral membrane (neutral sphingomyelinase) (EC:3.1.4.12) | NAD affects the activity of SMPD2 protein |
affects activity
|
protein |
20518072
|
| D009243 | 8877 |
SPHK1
SPHK |
sphingosine kinase 1 (EC:2.7.1.91) | NAD results in decreased activity of SPHK1 protein |
decreases activity
|
protein |
20518072
|
| D009243 | 7157 |
TP53
BCC7 LFS1 P53 TRP53 |
tumor protein p53 | [2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of TP53 gene |
affects cotreatment
/ increases mutagenesis |
gene |
10334203
|
| D009243 | 7157 |
TP53
BCC7 LFS1 P53 TRP53 |
tumor protein p53 | bathocuproine inhibits the reaction [[2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of TP53 gene] |
affects cotreatment
/ decreases reaction / increases mutagenesis |
gene |
10334203
|
| D009243 | 7157 |
TP53
BCC7 LFS1 P53 TRP53 |
tumor protein p53 | CAT protein inhibits the reaction [[2-phenylphenol metabolite co-treated with NAD co-treated with Copper] results in increased mutagenesis of TP53 gene] |
affects cotreatment
/ decreases reaction / increases mutagenesis |
gene |
10334203
|
| D009243 | 7157 |
TP53
BCC7 LFS1 P53 TRP53 |
tumor protein p53 | NAD results in decreased expression of TP53 protein |
decreases expression
|
protein |
12390773
|
| D009243 | 7299 |
TYR
ATN CMM8 OCA1 OCA1A OCAIA SHEP3 |
tyrosinase (EC:1.14.18.1) | ethylenediamine inhibits the reaction [[TYR protein results in increased oxidation of caffeic acid phenethyl ester] which results in increased oxidation of NAD] |
decreases reaction
/ increases oxidation |
protein |
20685355
|
| D009243 | 7299 |
TYR
ATN CMM8 OCA1 OCA1A OCAIA SHEP3 |
tyrosinase (EC:1.14.18.1) | Glutathione inhibits the reaction [[TYR protein results in increased oxidation of caffeic acid phenethyl ester] which results in increased oxidation of NAD] |
decreases reaction
/ increases oxidation |
protein |
20685355
|
| D009243 | 7299 |
TYR
ATN CMM8 OCA1 OCA1A OCAIA SHEP3 |
tyrosinase (EC:1.14.18.1) | [TYR protein results in increased oxidation of caffeic acid phenethyl ester] which results in increased oxidation of NAD |
increases oxidation
|
protein |
20685355
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (25)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #127750 | Dementia, lewy body; dlb |
P37840
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #612933 | Glycogen storage disease xi; gsd11 |
P00338
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #254210 | Myasthenic syndrome, congenital, associated with episodic apnea |
P28329
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #168601 | Parkinson disease 1, autosomal dominant; park1 |
P37840
|
| #605543 | Parkinson disease 4, autosomal dominant; park4 |
P37840
|
| #168600 | Parkinson disease, late-onset; pd |
P37840
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (24)
| KEGG | disease name | UniProt |
|---|---|---|
| H00069 | Glycogen storage diseases (GSD) |
P00338
(related)
|
| H00010 | Multiple myeloma |
P00338
(marker)
|
| H00023 | Testicular cancer |
P00338
(marker)
|
| H00035 | Ewing's sarcoma |
P00338
(marker)
|
| H00043 | Neuroblastoma |
P00338
(marker)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00770 | Congenital myasthenic syndrome |
P28329
(related)
|
| H00057 | Parkinson's disease (PD) |
P37840
(related)
|
| H00066 | Lewy body dementia (LBD) |
P37840
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
Q13148
(related)
|
Diseases related to CTD interactions
3 disease from interactions of metabolites
| MeSH disease | OMIM | compound | disease name | evidence type |
reference
pmid |
|---|---|---|---|---|---|
| C537948 | D009243 | Ceroid lipofuscinosis, neuronal 1, infantile |
marker/mechanism
|
21224254
|
|
| D056486 | D009243 | Drug-Induced Liver Injury |
therapeutic
|
11461765
|
|
| D009336 | D009243 | Necrosis |
marker/mechanism
|
19561459
|