KCF-S cluster No. 2752 (4 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| rosids | 1 |
Cumulative family count
| class name | count |
|---|---|
| Enterobacteriaceae | 4 |
| Brassicaceae | 1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (4)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00007313
|
UDP
/ Uridine 5'-diphosphate |
CHEMBL130266
CHEMBL1625810 |
D014530
|
14 / 6 / 6 | 4 / 0 |

|
|
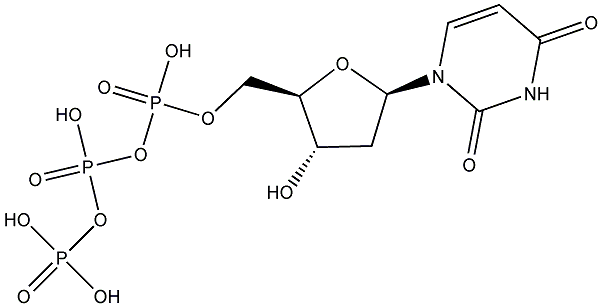
C00019636
|
dUTP
/ 2'-Deoxyuridine 5'-triphosphate |
CHEMBL374361
CHEMBL252573 |
C027078
|
1 / 0 / 0 |
|
|
|
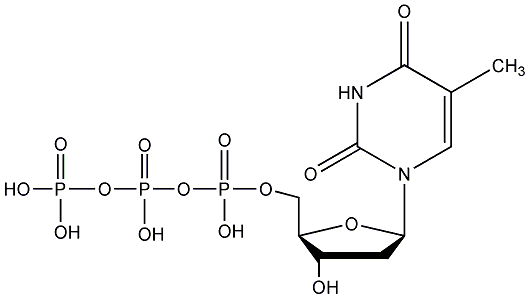
C00019695
|
TTP
/ dTTP / Deoxythymidine triphosphate / Deoxythymidine 5'-triphosphate |
CHEMBL363559
CHEMBL1674 |
C024157
|
|
||
|
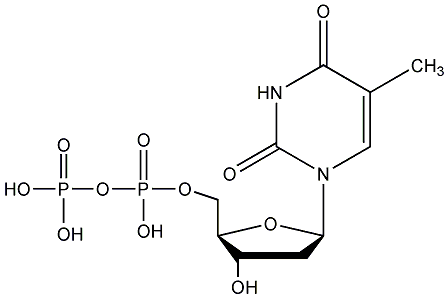
C00019696
|
dTDP
/ Thymidine 5'-diphosphate |
CHEMBL259724
|
C027923
|
2 / 0 / 0 |
|
Human Protein / Gene in interactions
16 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P41231 | P2Y purinoceptor 2 | Purine receptor | C00007313 C00019636 | 0 / 0 |
| Q15391 | P2Y purinoceptor 14 | Purine receptor | C00007313 | 0 / 0 |
| P51582 | P2Y purinoceptor 4 | Purine receptor | C00007313 | 0 / 0 |
| P22413 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | Enzyme | C00007313 | 4 / 4 |
| Q9Y271 | Cysteinyl leukotriene receptor 1 | Leukotriene receptor | C00007313 | 0 / 0 |
| P27708 | CAD protein | Enzyme | C00007313 | 0 / 0 |
| P47900 | P2Y purinoceptor 1 | Purine receptor | C00007313 | 0 / 0 |
| O14638 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | Enzyme | C00007313 | 0 / 0 |
| Q15077 | P2Y purinoceptor 6 | Purine receptor | C00007313 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00007313 | 0 / 0 |
| P15531 | Nucleoside diphosphate kinase A | Enzyme | C00019696 | 0 / 0 |
| P22392 | Nucleoside diphosphate kinase B | Enzyme | C00019696 | 0 / 0 |
| O15294 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | Enzyme | C00007313 | 0 / 0 |
| Q7LG56 | Ribonucleoside-diphosphate reductase subunit M2 B | Enzyme | C00007313 | 2 / 2 |
| P31350 | Ribonucleoside-diphosphate reductase subunit M2 | Enzyme | C00007313 | 0 / 0 |
| P23921 | Ribonucleoside-diphosphate reductase large subunit | Oxidoreductase | C00007313 | 0 / 0 |
4 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 10800 | CYSLTR1, CYSLT1, CYSLT1R, CYSLTR, HMTMF81 | cysteinyl leukotriene receptor 1 |
C00007313
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00007313
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00007313
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00007313
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (6)
| OMIM | preferred title | UniProt |
|---|---|---|
| #208000 | Arterial calcification, generalized, of infancy, 1; gaci1 |
P22413
|
| #125853 | Diabetes mellitus, noninsulin-dependent; niddm |
P22413
|
| #613312 | Hypophosphatemic rickets, autosomal recessive, 2; arhr2 |
P22413
|
| #612075 | Mitochondrial dna depletion syndrome 8a (encephalomyopathic type with renal tubulopathy); mtdps8a |
Q7LG56
|
| %602475 | Ossification of the posterior longitudinal ligament of spine; opll |
P22413
|
| #613077 | Progressive external ophthalmoplegia with mitochondrial dna deletions, autosomal dominant, 5; peoa5 |
Q7LG56
|
KEGG DISEASE (6)
| KEGG | name | UniProt |
|---|---|---|
| H00214 | Hypophosphatemic rickets |
P22413
(related)
|
| H00409 | Type II diabetes mellitus |
P22413
(related)
|
| H00431 | Ossification of the posterior longitudinal ligament of spine (OPLL) |
P22413
(related)
|
| H01002 | Generalized arterial calcification of infancy |
P22413
(related)
|
| H00469 | Mitochondrial DNA depletion syndrome (MDS) |
Q7LG56
(related)
|
| H01118 | Progressive external ophthalmoplegia (PEO) |
Q7LG56
(related)
|