KCF-S cluster No. 3630 (3 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|
Cumulative family count
| class name | count |
|---|---|
| Methanobacteriaceae | 2 |
| Methanosarcinaceae | 2 |
| Axinellidae | 1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (3)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00000761
|
Coenzyme M
|
CHEMBL1098319
|
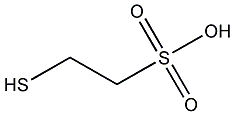
|
|||
|
C00000762
|
S-Methyl coenzyme M
|
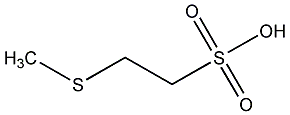
|
||||
|
C00048188
|
Taurine
|
CHEMBL239243
|
D013654
|
14 / 21 / 19 | 55 / 38 |
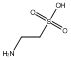
|
Human Protein / Gene in interactions
14 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00048188 | 1 / 0 |
| P04062 | Glucosylceramidase | Enzyme | C00048188 | 6 / 4 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00048188 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00048188 | 11 / 10 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00048188 | 0 / 1 |
| P54132 | Bloom syndrome protein | Enzyme | C00048188 | 1 / 2 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00048188 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00048188 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00048188 | 0 / 1 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00048188 | 0 / 0 |
| Q7Z2H8 | Proton-coupled amino acid transporter 1 | Unclassified protein | C00048188 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00048188 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00048188 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00048188 | 1 / 0 |
55 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 27 | ABL2, ABLL, ARG | c-abl oncogene 2, non-receptor tyrosine kinase (EC:2.7.10.2) |
C00048188
|
| 55803 | ADAP2, CENTA2, HSA272195, cent-b | ArfGAP with dual PH domains 2 |
C00048188
|
| 135 | ADORA2A, A2aR, ADORA2, RDC8 | adenosine A2a receptor |
C00048188
|
| 147 | ADRA1B, ADRA1, ALPHA1BAR | adrenoceptor alpha 1B |
C00048188
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00048188
|
| 659 | BMPR2, BMPR-II, BMPR3, BMR2, BRK-3, PPH1, T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) (EC:2.7.11.30) |
C00048188
|
| 10486 | CAP2 | CAP, adenylate cyclase-associated protein, 2 (yeast) |
C00048188
|
| 1081 | CGA, CG-ALPHA, FSHA, GPHA1, GPHa, HCG, LHA, TSHA | glycoprotein hormones, alpha polypeptide |
C00048188
|
| 1123 | CHN1, ARHGAP2, CHN, DURS2, NC, RHOGAP2 | chimerin 1 |
C00048188
|
| 9318 | COPS2, ALIEN, CSN2, SGN2, TRIP15 | COP9 signalosome subunit 2 |
C00048188
|
| 2833 | CXCR3, CD182, CD183, CKR-L2, CMKAR3, GPR9, IP10-R, Mig-R, MigR | chemokine (C-X-C motif) receptor 3 |
C00048188
|
| 23576 | DDAH1, DDAH | dimethylarginine dimethylaminohydrolase 1 (EC:3.5.3.18) |
C00048188
|
| 8525 | DGKZ, DAGK5, DAGK6, DGK-ZETA, hDGKzeta | diacylglycerol kinase, zeta (EC:2.7.1.107) |
C00048188
|
| 9538 | EI24, EPG4, PIG8, TP53I8 | etoposide induced 2.4 |
C00048188
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00048188
|
| 2260 | FGFR1, BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | fibroblast growth factor receptor 1 (EC:2.7.10.1) |
C00048188
|
| 2741 | GLRA1, HKPX1, STHE | glycine receptor, alpha 1 |
C00048188
|
| 2917 | GRM7, GLUR7, GPRC1G, MGLU7, MGLUR7 | glutamate receptor, metabotropic 7 |
C00048188
|
| 2982 | GUCY1A3, GC-SA3, GUC1A3, GUCA3, GUCSA3, GUCY1A1 | guanylate cyclase 1, soluble, alpha 3 (EC:4.6.1.2) |
C00048188
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00048188
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00048188
|
| 889 | KRIT1, CAM, CCM1 | KRIT1, ankyrin repeat containing |
C00048188
|
| 10314 | LANCL1, GPR69A, p40 | LanC lantibiotic synthetase component C-like 1 (bacterial) |
C00048188
|
| 4067 | LYN, JTK8, p53Lyn, p56Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (EC:2.7.10.2) |
C00048188
|
| 64231 | MS4A6A, 4SPAN3, 4SPAN3.2, CD20L3, MS4A6, MST090, MSTP090 | membrane-spanning 4-domains, subfamily A, member 6A |
C00048188
|
| 81285 | OR51E2, OR51E3P, OR52A2, PSGR | olfactory receptor, family 51, subfamily E, member 2 |
C00048188
|
| 55824 | PAG1, CBP, PAG | phosphoprotein associated with glycosphingolipid microdomains 1 |
C00048188
|
| 10298 | PAK4 | p21 protein (Cdc42/Rac)-activated kinase 4 (EC:2.7.11.1) |
C00048188
|
| 5423 | POLB | polymerase (DNA directed), beta (EC:2.7.7.7) |
C00048188
|
| 5424 | POLD1, CDC2, CRCS10, MDPL, POLD | polymerase (DNA directed), delta 1, catalytic subunit (EC:2.7.7.7) |
C00048188
|
| 27434 | POLM, Pol_Mu, Tdt-N | polymerase (DNA directed), mu (EC:2.7.7.7) |
C00048188
|
| 26051 | PPP1R16B, ANKRD4, TIMAP | protein phosphatase 1, regulatory subunit 16B |
C00048188
|
| 5529 | PPP2R5E | protein phosphatase 2, regulatory subunit B', epsilon isoform |
C00048188
|
| 5577 | PRKAR2B, PRKAR2, RII-BETA | protein kinase, cAMP-dependent, regulatory, type II, beta (EC:2.7.11.1) |
C00048188
|
| 5664 | PSEN2, AD3L, AD4, CMD1V, PS2, STM2 | presenilin 2 (Alzheimer disease 4) |
C00048188
|
| 5796 | PTPRK, R-PTP-kappa | protein tyrosine phosphatase, receptor type, K (EC:3.1.3.48) |
C00048188
|
| 5868 | RAB5A, RAB5 | RAB5A, member RAS oncogene family |
C00048188
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00048188
|
| 5883 | RAD9A, RAD9 | RAD9 homolog A (S. pombe) (EC:3.1.11.2) |
C00048188
|
| 9693 | RAPGEF2, CNrasGEF, NRAPGEP, PDZ-GEF1, PDZGEF1, RA-GEF, RA-GEF-1, Rap-GEP, nRap_GEP | Rap guanine nucleotide exchange factor (GEF) 2 |
C00048188
|
| 10125 | RASGRP1, CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
C00048188
|
| 6196 | RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-alpha2, p90-RSK3, pp90RSK3 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 (EC:2.7.11.1) |
C00048188
|
| 6447 | SCG5, 7B2, P7B2, SGNE1, SgV | secretogranin V (7B2 protein) |
C00048188
|
| 9126 | SMC3, BAM, BMH, CDLS3, CSPG6, HCAP, SMC3L1 | structural maintenance of chromosomes 3 |
C00048188
|
| 10302 | SNAPC5, SNAP19 | small nuclear RNA activating complex, polypeptide 5, 19kDa |
C00048188
|
| 10253 | SPRY2, hSPRY2 | sprouty homolog 2 (Drosophila) |
C00048188
|
| 6729 | SRP54 | signal recognition particle 54kDa |
C00048188
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00048188
|
| 6869 | TACR1, NK1R, NKIR, SPR, TAC1R | tachykinin receptor 1 |
C00048188
|
| 7010 | TEK, CD202B, TIE-2, TIE2, VMCM, VMCM1 | TEK tyrosine kinase, endothelial (EC:2.7.10.1) |
C00048188
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00048188
|
| 9540 | TP53I3, PIG3 | tumor protein p53 inducible protein 3 |
C00048188
|
| 7297 | TYK2, JTK1 | tyrosine kinase 2 (EC:2.7.10.2) |
C00048188
|
| 7415 | VCP, ALS14, IBMPFD, IBMPFD1, TERA, p97 | valosin containing protein (EC:3.6.4.6) |
C00048188
|
| 7518 | XRCC4 | X-ray repair complementing defective repair in Chinese hamster cells 4 |
C00048188
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (21)
| OMIM | preferred title | UniProt |
|---|---|---|
| #210900 | Bloom syndrome; blm |
P54132
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
KEGG DISEASE (19)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
Diseases related to CTD interactions
38 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D055371 | Acute Lung Injury |
C00048188
|
| D000860 | Anoxia |
C00048188
|
| D001145 | Arrhythmias, Cardiac |
C00048188
|
| D001919 | Bradycardia |
C00048188
|
| D009202 | Cardiomyopathies |
C00048188
|
| D003329 | Coronary Vasospasm |
C00048188
|
| D003921 | Diabetes Mellitus, Experimental |
C00048188
|
| D003922 | Diabetes Mellitus, Type 1 |
C00048188
|
| D058065 | Diabetic Cardiomyopathies |
C00048188
|
| D003928 | Diabetic Nephropathies |
C00048188
|
| D056486 | Drug-Induced Liver Injury |
C00048188
|
| D050171 | Dyslipidemias |
C00048188
|
| D004487 | Edema |
C00048188
|
| D019446 | Endotoxemia |
C00048188
|
| D005198 | Fanconi Syndrome |
C00048188
|
| D005355 | Fibrosis |
C00048188
|
| D005923 | Glomerulosclerosis, Focal Segmental |
C00048188
|
| D006323 | Heart Arrest |
C00048188
|
| D006331 | Heart Diseases |
C00048188
|
| D006470 | Hemorrhage |
C00048188
|
| D006943 | Hyperglycemia |
C00048188
|
| D017495 | Hyperpigmentation |
C00048188
|
| D006973 | Hypertension |
C00048188
|
| D006984 | Hypertrophy |
C00048188
|
| D007022 | Hypotension |
C00048188
|
| D007249 | Inflammation |
C00048188
|
| D007674 | Kidney Diseases |
C00048188
|
| D007859 | Learning Disorders |
C00048188
|
| D008106 | Liver Cirrhosis, Experimental |
C00048188
|
| D009069 | Movement Disorders |
C00048188
|
| D009133 | Muscular Atrophy |
C00048188
|
| D009203 | Myocardial Infarction |
C00048188
|
| D009336 | Necrosis |
C00048188
|
| D009845 | Oligospermia |
C00048188
|
| D011014 | Pneumonia |
C00048188
|
| D011507 | Proteinuria |
C00048188
|
| D012640 | Seizures |
C00048188
|
| D013733 | Testicular Diseases |
C00048188
|