KCF-S cluster No. 5945 (1 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| rosids | 3 |
| asterids | 1 |
| eudicotyledons | 1 |
Cumulative family count
| class name | count |
|---|---|
| Asteraceae | 1 |
| Combretaceae | 1 |
| Fabaceae | 1 |
| Ranunculaceae | 1 |
| Brassicaceae | 1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (1)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
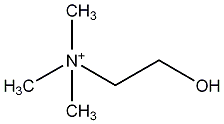
C00007298
|
Choline
|
CHEMBL920
|
D002794
|
11 / 5 / 5 | 51 / 20 |
|
Human Protein / Gene in interactions
11 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00007298 | 1 / 0 |
| O15244 | Solute carrier family 22 member 2 | Drug uniporter | C00007298 | 0 / 0 |
| O15245 | Solute carrier family 22 member 1 | Drug uniporter | C00007298 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00007298 | 0 / 1 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00007298 | 0 / 0 |
| P22303 | Acetylcholinesterase | Hydrolase | C00007298 | 1 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00007298 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00007298 | 0 / 1 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00007298 | 0 / 0 |
| Q9H015 | Solute carrier family 22 member 4 | Unclassified protein | C00007298 | 1 / 1 |
| O76082 | Solute carrier family 22 member 5 | Unclassified protein | C00007298 | 1 / 2 |
51 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 43 | ACHE, ACEE, ARACHE, N-ACHE, YT | acetylcholinesterase (EC:3.1.1.7) |
C00007298
|
| 8854 | ALDH1A2, RALDH(II), RALDH2, RALDH2-T | aldehyde dehydrogenase 1 family, member A2 (EC:1.2.1.36) |
C00007298
|
| 64333 | ARHGAP9, 10C, RGL1 | Rho GTPase activating protein 9 |
C00007298
|
| 590 | BCHE, CHE1, CHE2, E1 | butyrylcholinesterase (EC:3.1.1.8) |
C00007298
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00007298
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00007298
|
| 9332 | CD163, M130, MM130 | CD163 molecule |
C00007298
|
| 948 | CD36, BDPLT10, CHDS7, FAT, GP3B, GP4, GPIV, PASIV, SCARB3 | CD36 molecule (thrombospondin receptor) |
C00007298
|
| 55143 | CDCA8, BOR, BOREALIN, DasraB, MESRGP | cell division cycle associated 8 |
C00007298
|
| 55349 | CHDH | choline dehydrogenase (EC:1.1.99.1) |
C00007298
|
| 1111 | CHEK1, CHK1 | checkpoint kinase 1 (EC:2.7.11.1) |
C00007298
|
| 1137 | CHRNA4, BFNC, EBN, EBN1, NACHR, NACHRA4, NACRA4 | cholinergic receptor, nicotinic, alpha 4 (neuronal) |
C00007298
|
| 1139 | CHRNA7, CHRNA7-2, NACHRA7 | cholinergic receptor, nicotinic, alpha 7 (neuronal) |
C00007298
|
| 1141 | CHRNB2, EFNL3, nAChRB2 | cholinergic receptor, nicotinic, beta 2 (neuronal) |
C00007298
|
| 81035 | COLEC12, CLP1, NSR2, SCARA4, SRCL | collectin sub-family member 12 |
C00007298
|
| 1440 | CSF3, C17orf33, CSF3OS, GCSF, G-CSF | colony stimulating factor 3 (granulocyte) |
C00007298
|
| 1471 | CST3, ARMD11 | cystatin C |
C00007298
|
| 1514 | CTSL, CATL, CTSL1, MEP | cathepsin L (EC:3.4.22.15) |
C00007298
|
| 1524 | CX3CR1, CCRL1, CMKBRL1, CMKDR1, GPR13, GPRV28, V28 | chemokine (C-X3-C motif) receptor 1 |
C00007298
|
| 1794 | DOCK2 | dedicator of cytokinesis 2 |
C00007298
|
| 3169 | FOXA1, HNF3A, TCF3A | forkhead box A1 |
C00007298
|
| 2632 | GBE1, APBD, GBE, GSD4 | glucan (1,4-alpha-), branching enzyme 1 (EC:2.4.1.18) |
C00007298
|
| 3003 | GZMK, TRYP2 | granzyme K (granzyme 3; tryptase II) (EC:3.4.21.-) |
C00007298
|
| 3309 | HSPA5, BIP, GRP78, MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
C00007298
|
| 396742 |
C00007298
|
||
| 3560 | IL2RB, CD122, IL15RB, P70-75 | interleukin 2 receptor, beta |
C00007298
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00007298
|
| 9493 | KIF23, CHO1, KNSL5, MKLP-1, MKLP1 | kinesin family member 23 |
C00007298
|
| 51348 | KLRF1, CLEC5C, NKp80 | killer cell lectin-like receptor subfamily F, member 1 |
C00007298
|
| 114569 | MAL2 | mal, T-cell differentiation protein 2 (gene/pseudogene) |
C00007298
|
| 55016 | MARCH1, MARCH-I, RNF171 | membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
C00007298
|
| 4256 | MGP, MGLAP, NTI | matrix Gla protein |
C00007298
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00007298
|
| 4522 | MTHFD1, MTHFC, MTHFD | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase (EC:1.5.1.5 6.3.4.3 3.5.4.9) |
C00007298
|
| 9172 | MYOM2, TTNAP | myomesin 2 |
C00007298
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00007298
|
| 11333 | PDAP1, HASPP28, PAP, PAP1 | PDGFA associated protein 1 |
C00007298
|
| 10400 | PEMT, PEAMT, PEMPT, PEMT2, PNMT | phosphatidylethanolamine N-methyltransferase (EC:2.1.1.17 2.1.1.71) |
C00007298
|
| 5329 | PLAUR, CD87, U-PAR, UPAR, URKR | plasminogen activator, urokinase receptor |
C00007298
|
| 10891 | PPARGC1A, LEM6, PGC-1(alpha), PGC-1v, PGC1, PGC1A, PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
C00007298
|
| 23532 | PRAME, CT130, MAPE, OIP-4, OIP4 | preferentially expressed antigen in melanoma |
C00007298
|
| 5790 | PTPRCAP, CD45-AP, LPAP | protein tyrosine phosphatase, receptor type, C-associated protein |
C00007298
|
| 10287 | RGS19, GAIP, RGSGAIP | regulator of G-protein signaling 19 |
C00007298
|
| 57140 | RNPEPL1 | arginyl aminopeptidase (aminopeptidase B)-like 1 |
C00007298
|
| 6422 | SFRP1, FRP, FRP-1, FRP1, FrzA, SARP2 | secreted frizzled-related protein 1 |
C00007298
|
| 6584 | SLC22A5, CDSP, OCTN2, OCTN2VT | solute carrier family 22 (organic cation/carnitine transporter), member 5 |
C00007298
|
| 9056 | SLC7A7, LAT3, LPI, MOP-2, Y+LAT1, y+LAT-1 | solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
C00007298
|
| 6594 | SMARCA1, ISWI, NURF140, SNF2L, SNF2L1, SNF2LB, SNF2LT, SWI, SWI2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
C00007298
|
| 7015 | TERT, CMM9, DKCA2, DKCB4, EST2, PFBMFT1, TCS1, TP2, TRT, hEST2, hTRT | telomerase reverse transcriptase (EC:2.7.7.49) |
C00007298
|
| 7128 | TNFAIP3, A20, OTUD7C, TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 (EC:3.4.19.12) |
C00007298
|
| 83549 | UCK1, URK1 | uridine-cytidine kinase 1 (EC:2.7.1.48) |
C00007298
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
Diseases related to CTD interactions
20 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000647 | Amnesia |
C00007298
|
| D001321 | Autistic Disorder |
C00007298
|
| D006528 | Carcinoma, Hepatocellular |
C00007298
|
| D003072 | Cognition Disorders |
C00007298
|
| D003866 | Depressive Disorder |
C00007298
|
| D002658 | Developmental Disabilities |
C00007298
|
| D056486 | Drug-Induced Liver Injury |
C00007298
|
| D005234 | Fatty Liver |
C00007298
|
| D005235 | Fatty Liver, Alcoholic |
C00007298
|
| D006505 | Hepatitis |
C00007298
|
| D007249 | Inflammation |
C00007298
|
| D007859 | Learning Disorders |
C00007298
|
| D008103 | Liver Cirrhosis |
C00007298
|
| D008113 | Liver Neoplasms |
C00007298
|
| D008569 | Memory Disorders |
C00007298
|
| C541083 | Non-alcoholic Fatty Liver Disease |
C00007298
|
| D010195 | Pancreatitis |
C00007298
|
| D016137 | Spina Bifida Cystica |
C00007298
|
| D013226 | Status Epilepticus |
C00007298
|
| D052878 | Urolithiasis |
C00007298
|