KCF-S cluster No. 6732 (1 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|
Cumulative family count
| class name | count |
|---|---|
| Bovidae | 1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (1)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
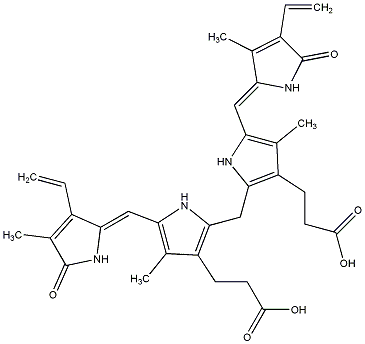
C00029828
|
Bilirubin
/ (Z,Z)-Bilirubin |
CHEMBL501680
|
D001663
|
22 / 31 / 28 | 155 / 20 |
|
Human Protein / Gene in interactions
22 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00029828 | 3 / 0 |
| O75795 | UDP-glucuronosyltransferase 2B17 | Enzyme | C00029828 | 1 / 0 |
| Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | Electrochemical transporter | C00029828 | 1 / 0 |
| P29466 | Caspase-1 | C14 | C00029828 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00029828 | 11 / 10 |
| P16662 | UDP-glucuronosyltransferase 2B7 | Enzyme | C00029828 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00029828 | 2 / 3 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00029828 | 0 / 0 |
| P46721 | Solute carrier organic anion transporter family member 1A2 | Unclassified protein | C00029828 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00029828 | 0 / 0 |
| Q9NPD5 | Solute carrier organic anion transporter family member 1B3 | Electrochemical transporter | C00029828 | 1 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00029828 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00029828 | 3 / 3 |
| P55210 | Caspase-7 | C14 | C00029828 | 0 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00029828 | 5 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00029828 | 4 / 3 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00029828 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00029828 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00029828 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00029828 | 1 / 1 |
| O00255 | Menin | Unclassified protein | C00029828 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00029828 | 1 / 2 |
155 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00029828
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00029828
|
| 210 | ALAD, ALADH, PBGS | aminolevulinate dehydratase (EC:4.2.1.24) |
C00029828
|
| 301 | ANXA1, ANX1, LPC1 | annexin A1 |
C00029828
|
| 374 | AREG, AR, CRDGF, SDGF | amphiregulin |
C00029828
|
| 9411 | ARHGAP29, PARG1, RP11-255E17.1 | Rho GTPase activating protein 29 |
C00029828
|
| 79754 | ASB13 | ankyrin repeat and SOCS box containing 13 |
C00029828
|
| 467 | ATF3 | activating transcription factor 3 |
C00029828
|
| 476 | ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (EC:3.6.3.9) |
C00029828
|
| 8313 | AXIN2, AXIL, ODCRCS | axin 2 |
C00029828
|
| 2647 | BLOC1S1, BLOS1, GCN5L1, MICoA, RT14 | biogenesis of lysosomal organelles complex-1, subunit 1 |
C00029828
|
| 10438 | C1D, LRP1, SUN-CoR, SUNCOR, hC1D | C1D nuclear receptor corepressor |
C00029828
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00029828
|
| 151887 | CCDC80, DRO1, SSG1, URB, okuribin | coiled-coil domain containing 80 |
C00029828
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00029828
|
| 25819 | CCRN4L, CCR4L, NOC | CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
C00029828
|
| 9936 | CD302, BIMLEC, CLEC13A, DCL-1, DCL1 | CD302 molecule |
C00029828
|
| 79094 | CHAC1 | ChaC, cation transport regulator homolog 1 (E. coli) |
C00029828
|
| 1119 | CHKA, CHK, CK, CKI, EK | choline kinase alpha (EC:2.7.1.32 2.7.1.82) |
C00029828
|
| 23155 | CLCC1, MCLC | chloride channel CLIC-like 1 |
C00029828
|
| 6249 | CLIP1, CLIP, CLIP-170, CLIP170, CYLN1, RSN | CAP-GLY domain containing linker protein 1 |
C00029828
|
| 1281 | COL3A1, EDS4A | collagen, type III, alpha 1 |
C00029828
|
| 1355 | COX15, CEMCOX2 | cytochrome c oxidase assembly homolog 15 (yeast) |
C00029828
|
| 1362 | CPD, GP180 | carboxypeptidase D (EC:3.4.17.22) |
C00029828
|
| 1437 | CSF2, GMCSF | colony stimulating factor 2 (granulocyte-macrophage) |
C00029828
|
| 1512 | CTSH, ACC-4, ACC-5, CPSB, minichain | cathepsin H (EC:3.4.22.16) |
C00029828
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00029828
|
| 6373 | CXCL11, H174, I-TAC, IP-9, IP9, SCYB11, SCYB9B, b-R1 | chemokine (C-X-C motif) ligand 11 |
C00029828
|
| 2920 | CXCL2, CINC-2a, GRO2, GROb, MGSA-b, MIP-2a, MIP2, MIP2A, SCYB2 | chemokine (C-X-C motif) ligand 2 |
C00029828
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00029828
|
| 1548 | CYP2A6, CPA6, CYP2A, CYP2A3, CYPIIA6, P450C2A, P450PB | cytochrome P450, family 2, subfamily A, polypeptide 6 (EC:1.14.14.1) |
C00029828
|
| 1555 | CYP2B6, CPB6, CYP2B, CYP2B7, CYP2B7P, CYPIIB6, EFVM, IIB1, P450 | cytochrome P450, family 2, subfamily B, polypeptide 6 (EC:1.14.14.1) |
C00029828
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00029828
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00029828
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00029828
|
| 420357 |
C00029828
|
||
| 93099 | DMKN, UNQ729, ZD52F10 | dermokine |
C00029828
|
| 3337 | DNAJB1, HSPF1, Hdj1, Hsp40, RSPH16B, Sis1 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
C00029828
|
| 23549 | DNPEP, ASPEP, DAP | aspartyl aminopeptidase (EC:3.4.11.21) |
C00029828
|
| 23333 | DPY19L1 | dpy-19-like 1 (C. elegans) |
C00029828
|
| 1847 | DUSP5, DUSP, HVH3 | dual specificity phosphatase 5 (EC:3.1.3.16 3.1.3.48) |
C00029828
|
| 54583 | EGLN1, C1orf12, ECYT3, HIF-PH2, HIFPH2, HPH-2, HPH2, PHD2, SM20, ZMYND6 | egl-9 family hypoxia-inducible factor 1 (EC:1.14.11.29) |
C00029828
|
| 9538 | EI24, EPG4, PIG8, TP53I8 | etoposide induced 2.4 |
C00029828
|
| 23741 | EID1, C15orf3, CRI1, EID-1, IRO45620, PTD014, RBP21 | EP300 interacting inhibitor of differentiation 1 |
C00029828
|
| 9451 | EIF2AK3, PEK, PERK, WRS | eukaryotic translation initiation factor 2-alpha kinase 3 (EC:2.7.11.1) |
C00029828
|
| 1965 | EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
C00029828
|
| 23052 | ENDOD1 | endonuclease domain containing 1 |
C00029828
|
| 345757 | FAM174A, TMEM157, UNQ1912 | family with sequence similarity 174, member A |
C00029828
|
| 8061 | FOSL1, FRA, FRA1, fra-1 | FOS-like antigen 1 |
C00029828
|
| 4616 | GADD45B, GADD45BETA, MYD118 | growth arrest and DNA-damage-inducible, beta |
C00029828
|
| 60674 | GAS5, NCRNA00030, SNHG2 | growth arrest-specific 5 (non-protein coding) |
C00029828
|
| 2934 | GSN, ADF, AGEL | gelsolin |
C00029828
|
| 2938 | GSTA1, GST2, GSTA1-1, GTH1 | glutathione S-transferase alpha 1 (EC:2.5.1.18) |
C00029828
|
| 2939 | GSTA2, GST2, GSTA2-2, GTA2, GTH2 | glutathione S-transferase alpha 2 (EC:2.5.1.18) |
C00029828
|
| 2940 | GSTA3, GSTA3-3, GTA3 | glutathione S-transferase alpha 3 (EC:2.5.1.18) |
C00029828
|
| 2941 | GSTA4, GSTA4-4, GTA4 | glutathione S-transferase alpha 4 (EC:2.5.1.18) |
C00029828
|
| 373156 | GSTK1, GST, GST_13-13, GST13, GST13-13, GSTK1-1, hGSTK1 | glutathione S-transferase kappa 1 (EC:2.5.1.18) |
C00029828
|
| 2944 | GSTM1, GST1, GSTM1-1, GSTM1a-1a, GSTM1b-1b, GTH4, GTM1, H-B, MU, MU-1 | glutathione S-transferase mu 1 (EC:2.5.1.18) |
C00029828
|
| 2950 | GSTP1, DFN7, FAEES3, GST3, GSTP, PI | glutathione S-transferase pi 1 (EC:2.5.1.18) |
C00029828
|
| 9843 | HEPH, CPL | hephaestin |
C00029828
|
| 9709 | HERPUD1, HERP, Mif1, SUP | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
C00029828
|
| 3008 | HIST1H1E, H1.4, H1E, H1F4, H1s-4, dJ221C16.5 | histone cluster 1, H1e |
C00029828
|
| 8347 | HIST1H2BC, H2B.1, H2B/l, H2BFL, dJ221C16.3 | histone cluster 1, H2bc |
C00029828
|
| 319188 |
C00029828
|
||
| 8294 | HIST1H4I, H4/m, H4FM, H4M | histone cluster 1, H4i |
C00029828
|
| 97114 |
C00029828
|
||
| 92815 | HIST3H2A | histone cluster 3, H2a |
C00029828
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00029828
|
| 373300 |
C00029828
|
||
| 3309 | HSPA5, BIP, GRP78, MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
C00029828
|
| 3418 | IDH2, D2HGA2, ICD-M, IDH, IDHM, IDP, IDPM, mNADP-IDH | isocitrate dehydrogenase 2 (NADP+), mitochondrial (EC:1.1.1.42) |
C00029828
|
| 3475 | IFRD1, PC4, TIS7 | interferon-related developmental regulator 1 |
C00029828
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00029828
|
| 3606 | IL18, IGIF, IL-18, IL-1g, IL1F4 | interleukin 18 (interferon-gamma-inducing factor) |
C00029828
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00029828
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00029828
|
| 3912 | LAMB1, CLM, LIS5 | laminin, beta 1 |
C00029828
|
| 3920 | LAMP2, CD107b, LAMP-2, LAMPB, LGP110 | lysosomal-associated membrane protein 2 |
C00029828
|
| 54741 | LEPROT, LEPR, OB-RGRP, OBRGRP, VPS55 | leptin receptor overlapping transcript |
C00029828
|
| 3976 | LIF, CDF, DIA, HILDA, MLPLI | leukemia inhibitory factor |
C00029828
|
| 4035 | LRP1, A2MR, APOER, APR, CD91, IGFBP3R, LRP, LRP1A, TGFBR5 | low density lipoprotein receptor-related protein 1 |
C00029828
|
| 110454 |
C00029828
|
||
| 4094 | MAF, CCA4, c-MAF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
C00029828
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00029828
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00029828
|
| 10982 | MAPRE2, EB1, EB2, RP1 | microtubule-associated protein, RP/EB family, member 2 |
C00029828
|
| 4200 | ME2, ODS1 | malic enzyme 2, NAD(+)-dependent, mitochondrial (EC:1.1.1.38) |
C00029828
|
| 1955 | MEGF9, EGFL5 | multiple EGF-like-domains 9 |
C00029828
|
| 4258 | MGST2, GST2, MGST-II | microsomal glutathione S-transferase 2 (EC:2.5.1.18) |
C00029828
|
| 4322 | MMP13, CLG3, MANDP1 | matrix metallopeptidase 13 (collagenase 3) (EC:3.4.24.-) |
C00029828
|
| 10205 | MPZL2, EVA, EVA1 | myelin protein zero-like 2 |
C00029828
|
| 29074 | MRPL18, L18mt, MRP-L18 | mitochondrial ribosomal protein L18 |
C00029828
|
| 4811 | NID1, NID | nidogen 1 |
C00029828
|
| 23530 | NNT, GCCD4 | nicotinamide nucleotide transhydrogenase (EC:1.6.1.2) |
C00029828
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00029828
|
| 9970 | NR1I3, CAR, CAR1, MB67 | nuclear receptor subfamily 1, group I, member 3 |
C00029828
|
| 29948 | OSGIN1, BDGI, OKL38 | oxidative stress induced growth inhibitor 1 |
C00029828
|
| 5163 | PDK1 | pyruvate dehydrogenase kinase, isozyme 1 (EC:2.7.11.2) |
C00029828
|
| 27315 | PGAP2, CWH43-N, FRAG1, HPMRS3, MRT17, MRT21 | post-GPI attachment to proteins 2 |
C00029828
|
| 5245 | PHB, PHB1 | prohibitin |
C00029828
|
| 8554 | PIAS1, DDXBP1, GBP, GU/RH-II, ZMIZ3 | protein inhibitor of activated STAT, 1 |
C00029828
|
| 5295 | PIK3R1, AGM7, GRB1, p85, p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
C00029828
|
| 18722 |
C00029828
|
||
| 51365 | PLA1A, PS-PLA1, PSPLA1 | phospholipase A1 member A (EC:3.1.1.-) |
C00029828
|
| 5376 | PMP22, CMT1A, CMT1E, DSS, GAS-3, HMSNIA, HNPP, Sp110 | peripheral myelin protein 22 |
C00029828
|
| 5445 | PON2 | paraoxonase 2 (EC:3.1.1.2 3.1.1.81) |
C00029828
|
| 10549 | PRDX4, AOE37-2, PRX-4 | peroxiredoxin 4 (EC:1.11.1.15) |
C00029828
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00029828
|
| 401494 | PTPLAD2, HACD4 | protein tyrosine phosphatase-like A domain containing 2 (EC:4.2.1.134) |
C00029828
|
| 83937 | RASSF4, AD037 | Ras association (RalGDS/AF-6) domain family member 4 |
C00029828
|
| 9584 | RBM39, CAPER, CAPERalpha, FSAP59, HCC1, RNPC2 | RNA binding motif protein 39 |
C00029828
|
| 10181 | RBM5, G15, LUCA15, RMB5 | RNA binding motif protein 5 |
C00029828
|
| 22836 | RHOBTB3 | Rho-related BTB domain containing 3 |
C00029828
|
| 6038 | RNASE4, RNS4 | ribonuclease, RNase A family, 4 (EC:3.1.27.-) |
C00029828
|
| 6281 | S100A10, 42C, ANX2L, ANX2LG, CAL1L, CLP11, Ca[1], GP11, P11, p10 | S100 calcium binding protein A10 |
C00029828
|
| 6275 | S100A4, 18A2, 42A, CAPL, FSP1, MTS1, P9KA, PEL98 | S100 calcium binding protein A4 |
C00029828
|
| 6342 | SCP2, NLTP, NSL-TP, SCP-2, SCP-CHI, SCP-X, SCPX | sterol carrier protein 2 (EC:2.3.1.176) |
C00029828
|
| 8991 | SELENBP1, LPSB, SBP56, SP56, hSBP | selenium binding protein 1 |
C00029828
|
| 66222 |
C00029828
|
||
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00029828
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00029828
|
| 81537 | SGPP1, SPPase1 | sphingosine-1-phosphate phosphatase 1 |
C00029828
|
| 387700 | SLC16A12, CJMG, MCT12 | solute carrier family 16, member 12 |
C00029828
|
| 9963 | SLC23A1, SLC23A2, SVCT1, YSPL3 | solute carrier family 23 (ascorbic acid transporter), member 1 |
C00029828
|
| 9962 | SLC23A2, NBTL1, SLC23A1, SVCT2, YSPL2 | solute carrier family 23 (ascorbic acid transporter), member 2 |
C00029828
|
| 6520 | SLC3A2, 4F2, 4F2HC, 4T2HC, CD98, CD98HC, MDU1, NACAE | solute carrier family 3 (amino acid transporter heavy chain), member 2 |
C00029828
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00029828
|
| 10599 | SLCO1B1, HBLRR, LST-1, LST1, OATP-C, OATP1B1, OATP2, OATPC, SLC21A6 | solute carrier organic anion transporter family, member 1B1 |
C00029828
|
| 57231 | SNX14, RGS-PX2 | sorting nexin 14 |
C00029828
|
| 50964 | SOST, CDD, VBCH | sclerostin |
C00029828
|
| 389058 | SP5 | Sp5 transcription factor |
C00029828
|
| 73526 |
C00029828
|
||
| 6892 | TAPBP, NGS17, TAPA, TPN, TPSN | TAP binding protein (tapasin) |
C00029828
|
| 6925 | TCF4, E2-2, ITF-2, ITF2, PTHS, SEF-2, SEF2, SEF2-1, SEF2-1A, SEF2-1B, TCF-4, bHLHb19 | transcription factor 4 |
C00029828
|
| 25976 | TIPARP, ARTD14, PARP7, pART14 | TCDD-inducible poly(ADP-ribose) polymerase (EC:2.4.2.30) |
C00029828
|
| 9375 | TM9SF2, P76 | transmembrane 9 superfamily member 2 |
C00029828
|
| 55504 | TNFRSF19, TAJ, TAJ-alpha, TRADE, TROY | tumor necrosis factor receptor superfamily, member 19 |
C00029828
|
| 1200 | TPP1, CLN2, LPIC, TPP-1 | tripeptidyl peptidase I (EC:3.4.14.9) |
C00029828
|
| 28951 | TRIB2, C5FW, GS3955, TRB2 | tribbles pseudokinase 2 |
C00029828
|
| 7329 | UBE2I, C358B7.1, P18, UBC9 | ubiquitin-conjugating enzyme E2I (EC:6.3.2.19) |
C00029828
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00029828
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00029828
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00029828
|
| 708396 |
C00029828
|
||
| 7381 | UQCRB, MC3DN3, QCR7, QP-C, QPC, UQBC, UQBP, UQCR6, UQPC | ubiquinol-cytochrome c reductase binding protein (EC:1.10.2.2) |
C00029828
|
| 7384 | UQCRC1, D3S3191, QCR1, UQCR1 | ubiquinol-cytochrome c reductase core protein I (EC:1.10.2.2) |
C00029828
|
| 7388 | UQCRH, QCR6, UQCR8 | ubiquinol-cytochrome c reductase hinge protein (EC:1.10.2.2) |
C00029828
|
| 80146 | UXS1, SDR6E1, UGD | UDP-glucuronate decarboxylase 1 (EC:4.1.1.35) |
C00029828
|
| 7436 | VLDLR, CARMQ1, CHRMQ1, VLDLRCH | very low density lipoprotein receptor |
C00029828
|
| 26137 | ZBTB20, DPZF, HOF, ODA-8S, ZNF288 | zinc finger and BTB domain containing 20 |
C00029828
|
| 339487 | ZBTB8OS, ARCH, ARCH2 | zinc finger and BTB domain containing 8 opposite strand |
C00029828
|
| 84186 | ZCCHC7, AIR1, RP11-397D12.1 | zinc finger, CCHC domain containing 7 |
C00029828
|
| 90637 | ZFAND2A, AIRAP | zinc finger, AN1-type domain 2A |
C00029828
|
| 22680 |
C00029828
|
||
| 24132 |
C00029828
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (31)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #612560 | Bone mineral density quantitative trait locus 12; bmnd12 |
O75795
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237450 | Hyperbilirubinemia, rotor type; hblrr |
Q9NPD5
Q9Y6L6 |
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (28)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q03164
(related)
Q03164 (marker) |
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q9NUW8
(related)
|
Diseases related to CTD interactions
20 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000743 | Anemia, Hemolytic |
C00029828
|
| D001145 | Arrhythmias, Cardiac |
C00029828
|
| D054537 | Atrioventricular Block |
C00029828
|
| D002779 | Cholestasis |
C00029828
|
| D002780 | Cholestasis, Intrahepatic |
C00029828
|
| D003244 | Consciousness Disorders |
C00029828
|
| D056486 | Drug-Induced Liver Injury |
C00029828
|
| D006504 | Hepatic Veno-Occlusive Disease |
C00029828
|
| D006505 | Hepatitis |
C00029828
|
| D006932 | Hyperbilirubinemia |
C00029828
|
| D006975 | Hypertension, Portal |
C00029828
|
| D007249 | Inflammation |
C00029828
|
| D007567 | Jaundice, Neonatal |
C00029828
|
| D007681 | Kidney Papillary Necrosis |
C00029828
|
| D008103 | Liver Cirrhosis |
C00029828
|
| D008107 | Liver Diseases |
C00029828
|
| D017093 | Liver Failure |
C00029828
|
| D009336 | Necrosis |
C00029828
|
| D012640 | Seizures |
C00029828
|
| D013226 | Status Epilepticus |
C00029828
|