Gratiola officinalis
Species
KNApSAcK Entry
| Organism name | Gratiola officinalis |
|---|---|
| Genus | Gratiola |
| Family | Plantaginaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Gratiola officinalis |
|---|---|
| Linked NCBI taxonomy ID | 204382 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Plantaginaceae |
|---|---|
| ID | 156152 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Natural Activity
List (10)
| Species | Activity |
|---|---|
| Gratiola officinalis L. | Cardiotonic |
| Gratiola officinalis L. | Cytotoxic |
| Gratiola officinalis L. | Digitalic |
| Gratiola officinalis L. | Diuretic |
| Gratiola officinalis L. | Emetic |
| Gratiola officinalis L. | Laxative |
| Gratiola officinalis L. | Parasiticide |
| Gratiola officinalis L. | toxic |
| Gratiola officinalis L. | Vermifuge |
| Gratiola officinalis L. | Vulnerary |
Metabolite list (7)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00013642
|
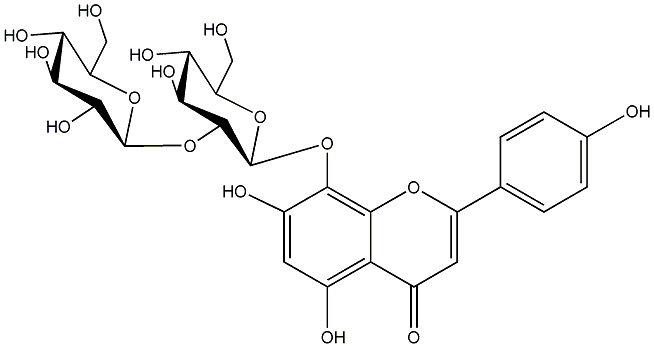
8-Hydroxyapigenin 8-sophoroside
/ 8-[(2-O-beta-D-Glucopyranosyl-beta-D-glucopyranosyl)oxy]-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
No. 1 | No. 15 |
|
||||
|
C00004497
|
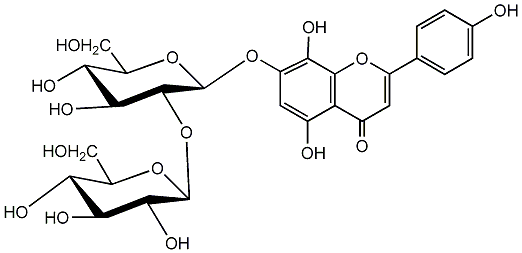
8-Hydroxyapigenin 7-sophoroside
|
No. 1 | No. 15 |
|
||||
|
C00004518
|
Hypolaetin 7-sophoroside
/ 8-Hydroxyluteolin 7-sophoroside / 7-[(2-O-beta-D-Glucopyranosyl-beta-D-glucopyranosyl)oxy]-2-(3,4-dihydroxyphenyl)-5,8-dihydroxy-4H-1-benzopyran-4-one |
No. 1 | No. 15 |

|
||||
|
C00004521
|
8-Hydroxyluteolin 4'-methyl ether 7-(6'''-acetylallosyl)(1->2)(6''-acetylglucoside)
|
No. 1 | No. 15 |

|
||||
|
C00004520
|
8-Hydroxyluteolin 3'-methyl ether 7-sophoroside
|
No. 1 | No. 15 |

|
||||
|
C00004519
|
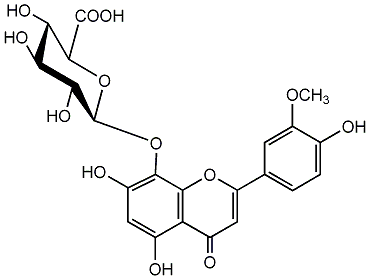
8-Hydroxychrysoeriol 8-glucuronide
/ 8-Hydroxyluteolin 3'-methyl ether 8-glucuronide / 5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4-oxo-4H-1-benzopyran-8-yl beta-D-glucopyranosiduronic acid |
No. 2 | No. 15 |
|
||||
|
C00003689
|
Cucurbitacin I
|
CHEMBL387737
CHEMBL1317135 CHEMBL1717364 |
C038106
|
20 / 15 / 15 | 2 / 0 | No. 220 | No. 11 |

|
Human Protein / Gene in interactions
20 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00003689 | 1 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00003689 | 4 / 1 |
| Q99700 | Ataxin-2 | Unclassified protein | C00003689 | 1 / 1 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003689 | 0 / 1 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00003689 | 2 / 3 |
| P20701 | Integrin alpha-L | Membrane receptor | C00003689 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00003689 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00003689 | 2 / 0 |
| O75496 | Geminin | Unclassified protein | C00003689 | 0 / 0 |
| P23528 | Cofilin-1 | Unclassified protein | C00003689 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00003689 | 0 / 0 |
| P11308 | Transcriptional regulator ERG | Unclassified protein | C00003689 | 1 / 2 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00003689 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003689 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00003689 | 0 / 1 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00003689 | 0 / 0 |
| P31939 | Bifunctional purine biosynthesis protein PURH | Enzyme | C00003689 | 1 / 1 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00003689 | 1 / 0 |
| P01215 | Glycoprotein hormones alpha chain | Unclassified protein | C00003689 | 0 / 3 |
| Q06710 | Paired box protein Pax-8 | Unclassified protein | C00003689 | 1 / 2 |
2 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 3717 | JAK2, JTK10, THCYT3 | Janus kinase 2 (EC:2.7.10.2) |
C00003689
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00003689
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (15)
| OMIM | preferred title | UniProt |
|---|---|---|
| #608688 | Aicar transformylase/imp cyclohydrolase deficiency |
P31939
|
| #114500 | Colorectal cancer; crc |
P84022
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #612219 | Ewing sarcoma; es |
P11308
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #218700 | Hypothyroidism, congenital, nongoitrous, 2; chng2 |
Q06710
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (15)
| KEGG | name | UniProt |
|---|---|---|
| H00081 | Hashimoto's thyroiditis |
P01215
(marker)
|
| H00082 | Graves' disease |
P01215
(marker)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P01215
(marker)
Q06710 (related) |
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00024 | Prostate cancer |
P11308
(related)
|
| H00035 | Ewing's sarcoma |
P11308
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00966 | AICA-ribosiduria |
P31939
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00032 | Thyroid cancer |
Q06710
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|