Streptomyces hygroscopicus var. enhygrus var. nov.
Species
KNApSAcK Entry
| Organism name | Streptomyces hygroscopicus var. enhygrus var. nov. |
|---|---|
| Genus | Streptomyces |
| Family | Streptomycetaceae |
| Kingdom | Bacteria |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Streptomyces |
|---|---|
| Linked NCBI taxonomy ID | 1883 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Streptomycetaceae |
|---|---|
| ID | 2062 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Bacteria |
|---|---|
| ID | 2 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (2)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00018829
|
U 29479
/ Nificin / NSC 107041 / Scopafungin / Niphimycin Ialpha |
CHEMBL413402
CHEMBL2103935 |
C000770
|
No. 383 |
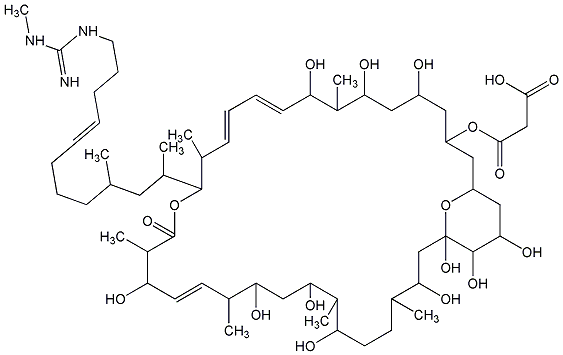
|
|||
|
C00018795
|
NSC 212518
/ NSC 122750 / Geldanamycin |
CHEMBL278315
CHEMBL182055 CHEMBL1165003 CHEMBL1594212 |
C001277
|
31 / 8 / 7 | 48 / 4 | No. 2569 |

|
Human Protein / Gene in interactions
31 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00018795 | 1 / 0 |
| Q9P1W9 | Serine/threonine-protein kinase pim-2 | Pim | C00018795 | 0 / 0 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00018795 | 2 / 2 |
| Q15759 | Mitogen-activated protein kinase 11 | p38 | C00018795 | 0 / 0 |
| Q9H2G2 | STE20-like serine/threonine-protein kinase | STE serine/threonine protein kinase SLK subfamily | C00018795 | 0 / 0 |
| Q16659 | Mitogen-activated protein kinase 6 | CMGC serine/threonine protein kinase ERK3 subfamily | C00018795 | 0 / 0 |
| Q16543 | Hsp90 co-chaperone Cdc37 | Unclassified protein | C00018795 | 0 / 0 |
| P11309 | Serine/threonine-protein kinase pim-1 | Pim | C00018795 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00018795 | 0 / 1 |
| O00444 | Serine/threonine-protein kinase PLK4 | PLK serine/threonine protein kinase subfamily | C00018795 | 0 / 0 |
| Q9NQU5 | Serine/threonine-protein kinase PAK 6 | STE serine/threonine protein kinase PAKB subfamily | C00018795 | 0 / 0 |
| O75828 | Carbonyl reductase [NADPH] 3 | Enzyme | C00018795 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00018795 | 2 / 3 |
| Q13188 | Serine/threonine-protein kinase 3 | STE serine/threonine protein kinase MST subfamily | C00018795 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00018795 | 0 / 0 |
| P07332 | Tyrosine-protein kinase Fes/Fps | Fer | C00018795 | 0 / 0 |
| P16152 | Carbonyl reductase [NADPH] 1 | Enzyme | C00018795 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00018795 | 2 / 0 |
| Q99683 | Mitogen-activated protein kinase kinase kinase 5 | Ste11 | C00018795 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00018795 | 0 / 0 |
| P52564 | Dual specificity mitogen-activated protein kinase kinase 6 | Ste7 | C00018795 | 0 / 0 |
| O14976 | Cyclin-G-associated kinase | NAK serine/threonine protein kinase subfamily | C00018795 | 0 / 0 |
| O94804 | Serine/threonine-protein kinase 10 | STE serine/threonine protein kinase SLK subfamily | C00018795 | 1 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00018795 | 0 / 0 |
| P08238 | Heat shock protein HSP 90-beta | Other cytosolic protein | C00018795 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00018795 | 0 / 1 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00018795 | 0 / 0 |
| Q9HCP0 | Casein kinase I isoform gamma-1 | Ck1 | C00018795 | 0 / 0 |
| P07900 | Heat shock protein HSP 90-alpha | Other cytosolic protein | C00018795 | 0 / 0 |
| Q86V86 | Serine/threonine-protein kinase pim-3 | Pim | C00018795 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00018795 | 0 / 0 |
48 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00018795
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00018795
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00018795
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00018795
|
| 329 | BIRC2, API1, HIAP2, Hiap-2, MIHB, RNF48, c-IAP1, cIAP1 | baculoviral IAP repeat containing 2 |
C00018795
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00018795
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00018795
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00018795
|
| 11140 | CDC37, P50CDC37 | cell division cycle 37 |
C00018795
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00018795
|
| 1080 | CFTR, ABC35, ABCC7, CF, CFTR/MRP, MRP7, TNR-CFTR, dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) (EC:3.6.3.49) |
C00018795
|
| 1147 | CHUK, IKBKA, IKK-alpha, IKK1, IKKA, NFKBIKA, TCF16 | conserved helix-loop-helix ubiquitous kinase (EC:2.7.11.10) |
C00018795
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00018795
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00018795
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00018795
|
| 2064 | ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NGL, TKR1 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (EC:2.7.10.1) |
C00018795
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00018795
|
| 2252 | FGF7, HBGF-7, KGF | fibroblast growth factor 7 |
C00018795
|
| 3091 | HIF1A, HIF-1A, HIF-1alpha, HIF1, HIF1-ALPHA, MOP1, PASD8, bHLHe78 | hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
C00018795
|
| 3248 | HPGD, 15-PGDH, PGDH, PGDH1, PHOAR1, SDR36C1 | hydroxyprostaglandin dehydrogenase 15-(NAD) (EC:1.1.1.141) |
C00018795
|
| 3297 | HSF1, HSTF1 | heat shock transcription factor 1 |
C00018795
|
| 692420 |
C00018795
|
||
| 3320 | HSP90AA1, EL52, HSP86, HSP89A, HSP90A, HSP90N, HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 |
C00018795
|
| 7184 | HSP90B1, ECGP, GP96, GRP94, TRA1 | heat shock protein 90kDa beta (Grp94), member 1 |
C00018795
|
| 3303 | HSPA1A, HSP70-1, HSP70-1A, HSP70I, HSP72, HSPA1 | heat shock 70kDa protein 1A |
C00018795
|
| 3308 | HSPA4, APG-2, HS24/P52, HSPH2, RY, hsp70, hsp70RY | heat shock 70kDa protein 4 |
C00018795
|
| 3309 | HSPA5, BIP, GRP78, MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
C00018795
|
| 3064 | HTT, HD, IT15 | huntingtin |
C00018795
|
| 11009 | IL24, C49A, FISP, IL10B, MDA7, MOB5, ST16 | interleukin 24 |
C00018795
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00018795
|
| 3757 | KCNH2, ERG1, HERG, HERG1, Kv11.1, LQT2, SQT1 | potassium voltage-gated channel, subfamily H (eag-related), member 2 |
C00018795
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00018795
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00018795
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00018795
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00018795
|
| 4255 | MGMT | O-6-methylguanine-DNA methyltransferase (EC:2.1.1.63) |
C00018795
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00018795
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00018795
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00018795
|
| 5241 | PGR, NR3C3, PR | progesterone receptor |
C00018795
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00018795
|
| 5744 | PTHLH, BDE2, HHM, PLP, PTHR, PTHRP | parathyroid hormone-like hormone |
C00018795
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00018795
|
| 5894 | RAF1, CRAF, NS5, Raf-1, c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 (EC:2.7.11.1) |
C00018795
|
| 6714 | SRC, ASV, SRC1, c-SRC, p60-Src | v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (EC:2.7.10.2) |
C00018795
|
| 10273 | STUB1, CHIP, HSPABP2, NY-CO-7, SDCCAG7, UBOX1 | STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase |
C00018795
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00018795
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00018795
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (8)
| OMIM | preferred title | UniProt |
|---|---|---|
| #114500 | Colorectal cancer; crc |
P84022
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #273300 | Testicular germ cell tumor; tgct |
O94804
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (7)
| KEGG | name | UniProt |
|---|---|---|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|