Angelica dahurica
Species
KNApSAcK Entry
| Organism name | Angelica dahurica |
|---|---|
| Genus | Angelica |
| Family | Apiaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Angelica dahurica |
|---|---|
| Linked NCBI taxonomy ID | 48101 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Apiaceae |
|---|---|
| ID | 4037 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (3)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00030532
|
Isoimperatorin
|
CHEMBL448060
|
C055542
|
8 / 13 / 11 | 3 / 1 | No. 606 | No. 25 |
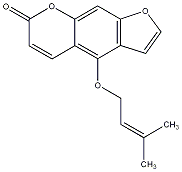
|
|
C00002477
|
Imperatorin
|
CHEMBL453805
|
C031534
|
18 / 7 / 6 | 1 / 1 | No. 606 | No. 25 |
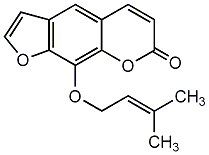
|
|
C00001283
|
Falcarindiol
|
CHEMBL69018
CHEMBL1094112 |
C034379
|
3 / 0 / 0 | No. 940 | No. 69 |
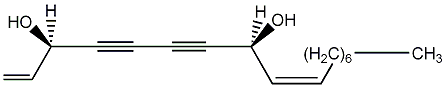
|
Human Protein / Gene in interactions
24 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00002477 C00030532 | 1 / 1 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002477 C00030532 | 0 / 0 |
| P56817 | Beta-secretase 1 | A1A | C00002477 C00030532 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002477 C00030532 | 0 / 0 |
| P34969 | 5-hydroxytryptamine receptor 7 | Serotonin receptor | C00001283 C00002477 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00002477 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00002477 | 2 / 3 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00001283 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00001283 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00030532 | 11 / 10 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00002477 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002477 | 0 / 0 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00002477 | 0 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00002477 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00002477 | 1 / 1 |
| P22303 | Acetylcholinesterase | Hydrolase | C00030532 | 1 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002477 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00030532 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00002477 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00002477 | 0 / 0 |
| O00167 | Eyes absent homolog 2 | Enzyme | C00002477 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00002477 | 2 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00002477 | 1 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00030532 | 0 / 0 |
3 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00002477
C00030532
|
| 2939 | GSTA2, GST2, GSTA2-2, GTA2, GTH2 | glutathione S-transferase alpha 2 (EC:2.5.1.18) |
C00030532
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00030532
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (19)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (16)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
Q13148
(related)
|