Botrytis cinerea
Species
KNApSAcK Entry
| Organism name | Botrytis cinerea |
|---|---|
| Genus | |
| Family | |
| Kingdom |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Botryotinia fuckeliana |
|---|---|
| Linked NCBI taxonomy ID | 40559 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Sclerotiniaceae |
|---|---|
| ID | 28983 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Fungi |
|---|---|
| ID | 4751 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (16)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00023751
|
Fecosterol
/ Ergosta-8,24(28)-dien-3beta-ol / 24-Methylcholesta-8,24(28)-dien-3beta-ol |
C039241
|
No. 111 | No. 11 |

|
|||
|
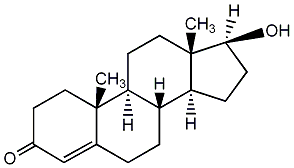
C00047082
|
Pregnenolone
|
CHEMBL77462
CHEMBL383839 CHEMBL253363 CHEMBL1435695 CHEMBL1455777 |
D011284
|
22 / 28 / 19 | 9 / 0 | No. 1223 |
|
|
|
C00021572
|
Botryoloic acid
|
CHEMBL478927
|
No. 1269 |

|
||||
|
C00021569
|
Botryaloic acid acetate
|
No. 1269 |

|
|||||
|
C00021568
|
Botryaloic acid
|
No. 1269 |

|
|||||
|
C00021573
|
Dihydrobotrydial
|
CHEMBL515958
CHEMBL477747 |
No. 1269 |

|
||||
|
C00046820
|
Methyl botryoloate
/ (+)-Methyl botryoloate |
CHEMBL480865
|
No. 1269 |

|
||||
|
C00046720
|
Dihydrobotrydialone
|
No. 1269 |

|
|||||
|
C00003104
|
Botrydial
|
No. 1269 |
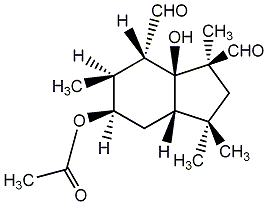
|
|||||
|
C00046209
|
Norbotrydialone acetate
/ (-)-Norbotrydialone acetate |
No. 1433 |

|
|||||
|
C00045507
|
10-Oxodehydrodihydrobotrydial
/ (+)-10-Oxodehydrodihydrobotrydial |
No. 1808 |

|
|||||
|
C00046716
|
Dehydrobotrydienol
/ (-)-Dehydrobotrydienol |
CHEMBL480575
|
No. 2191 |

|
||||
|
C00046557
|
12-Hydroxydehydrobotrydienol
/ (+)-12-Hydroxydehydrobotrydienol |
CHEMBL477706
|
No. 2191 |

|
||||
|
C00046551
|
11-Hydroxydehydrobotrydienol
/ (-)-11-Hydroxydehydrobotrydienol |
CHEMBL445801
|
No. 2191 |

|
||||
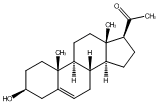
|
C00003675
|
Testosterone
|
CHEMBL268313
CHEMBL67934 CHEMBL196228 CHEMBL386630 CHEMBL1362515 |
D013739
|
145 / 100 / 73 | 1240 / 93 | No. 2299 | No. 54 |
|
|
C00046922
|
Secobotrytriendiol
/ (+)-Secobotrytriendiol |
CHEMBL480866
|
No. 5920 |

|
Human Protein / Gene in interactions
150 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00003675 C00047082 | 1 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00003675 C00047082 | 0 / 0 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00003675 C00047082 | 3 / 0 |
| O75795 | UDP-glucuronosyltransferase 2B17 | Enzyme | C00003675 C00047082 | 1 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00003675 C00047082 | 0 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003675 C00047082 | 1 / 1 |
| P08185 | Corticosteroid-binding globulin | Secreted protein | C00003675 C00047082 | 1 / 1 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00003675 C00047082 | 0 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00003675 C00047082 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00003675 C00047082 | 0 / 0 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00003675 C00047082 | 7 / 3 |
| P04278 | Sex hormone-binding globulin | Secreted protein | C00003675 C00047082 | 0 / 0 |
| Q06520 | Bile salt sulfotransferase | Enzyme | C00003675 C00047082 | 0 / 0 |
| P16662 | UDP-glucuronosyltransferase 2B7 | Enzyme | C00003675 C00047082 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003675 C00047082 | 0 / 1 |
| Q16850 | Lanosterol 14-alpha demethylase | Cytochrome P450 51A1 | C00003675 C00047082 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00003675 C00047082 | 11 / 10 |
| P36537 | UDP-glucuronosyltransferase 2B10 | Enzyme | C00003675 | 0 / 0 |
| P29466 | Caspase-1 | C14 | C00003675 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00003675 | 0 / 0 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00003675 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00003675 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00003675 | 2 / 2 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00003675 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00003675 | 0 / 0 |
| P49146 | Neuropeptide Y receptor type 2 | Neuropeptide Y receptor | C00003675 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00003675 | 3 / 2 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00003675 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00003675 | 1 / 2 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00003675 | 0 / 1 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00003675 | 1 / 1 |
| P25021 | Histamine H2 receptor | Histamine receptor | C00003675 | 0 / 0 |
| P35367 | Histamine H1 receptor | Histamine receptor | C00003675 | 0 / 0 |
| Q01959 | Sodium-dependent dopamine transporter | Dopamine | C00003675 | 1 / 0 |
| P08912 | Muscarinic acetylcholine receptor M5 | Acetylcholine receptor | C00003675 | 0 / 0 |
| P18825 | Alpha-2C adrenergic receptor | Adrenergic receptor | C00003675 | 0 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00003675 | 0 / 0 |
| P02768 | Serum albumin | Secreted protein | C00003675 | 0 / 0 |
| Q16539 | Mitogen-activated protein kinase 14 | p38 | C00003675 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00003675 | 1 / 0 |
| P25024 | C-X-C chemokine receptor type 1 | CXC chemokine receptor | C00003675 | 0 / 0 |
| P06241 | Tyrosine-protein kinase Fyn | Src | C00003675 | 0 / 0 |
| Q08209 | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ser_Thr | C00003675 | 0 / 0 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00003675 | 0 / 0 |
| P08246 | Neutrophil elastase | S1A | C00003675 | 2 / 1 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00003675 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00003675 | 1 / 8 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00003675 | 2 / 3 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | C00003675 | 2 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00003675 | 0 / 0 |
| P37288 | Vasopressin V1a receptor | Vasopressin and oxytocin receptor | C00003675 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00003675 | 0 / 0 |
| Q9Y271 | Cysteinyl leukotriene receptor 1 | Leukotriene receptor | C00003675 | 0 / 0 |
| P29274 | Adenosine receptor A2a | Adenosine receptor | C00003675 | 0 / 0 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00003675 | 2 / 2 |
| P25929 | Neuropeptide Y receptor type 1 | Neuropeptide Y receptor | C00003675 | 0 / 0 |
| P50052 | Type-2 angiotensin II receptor | Angiotensin receptor | C00003675 | 1 / 1 |
| P17948 | Vascular endothelial growth factor receptor 1 | Vegfr | C00003675 | 0 / 0 |
| P41968 | Melanocortin receptor 3 | Melanocortin receptor | C00003675 | 1 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00003675 | 0 / 3 |
| P11509 | Cytochrome P450 2A6 | Cytochrome P450 2A6 | C00003675 | 0 / 0 |
| P11413 | Glucose-6-phosphate 1-dehydrogenase | Enzyme | C00047082 | 1 / 2 |
| O75496 | Geminin | Unclassified protein | C00003675 | 0 / 0 |
| P54855 | UDP-glucuronosyltransferase 2B15 | Enzyme | C00003675 | 0 / 0 |
| P04035 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Oxidoreductase | C00003675 | 0 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00003675 | 0 / 0 |
| P21917 | D(4) dopamine receptor | Dopamine receptor | C00003675 | 0 / 0 |
| P30988 | Calcitonin receptor | Calcitonin receptor | C00003675 | 0 / 0 |
| P35462 | D(3) dopamine receptor | Dopamine receptor | C00003675 | 1 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00003675 | 0 / 0 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00003675 | 0 / 1 |
| P41595 | 5-hydroxytryptamine receptor 2B | Serotonin receptor | C00003675 | 0 / 0 |
| P25101 | Endothelin-1 receptor | Endothelin receptor | C00003675 | 0 / 0 |
| P30411 | B2 bradykinin receptor | Bradykinin receptor | C00003675 | 0 / 0 |
| P10145 | Interleukin-8 | Secreted protein | C00003675 | 0 / 0 |
| P32245 | Melanocortin receptor 4 | Melanocortin receptor | C00003675 | 1 / 0 |
| P32238 | Cholecystokinin receptor type A | Cholecystokinin receptor | C00003675 | 0 / 0 |
| P08311 | Cathepsin G | S1A | C00003675 | 0 / 0 |
| Q99720 | Sigma non-opioid intracellular receptor 1 | Membrane receptor | C00003675 | 1 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00003675 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00003675 | 0 / 1 |
| P32241 | Vasoactive intestinal polypeptide receptor 1 | Vasoactive intestinal peptide receptor | C00003675 | 0 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00003675 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00003675 | 0 / 0 |
| Q9NPD5 | Solute carrier organic anion transporter family member 1B3 | Electrochemical transporter | C00003675 | 1 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00003675 | 0 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00003675 | 0 / 1 |
| P06401 | Progesterone receptor | NR3C3 | C00003675 | 0 / 1 |
| P08172 | Muscarinic acetylcholine receptor M2 | Acetylcholine receptor | C00003675 | 2 / 0 |
| P11229 | Muscarinic acetylcholine receptor M1 | Acetylcholine receptor | C00003675 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00003675 | 2 / 2 |
| P21554 | Cannabinoid receptor 1 | Cannabinoid receptor | C00003675 | 0 / 0 |
| P31645 | Sodium-dependent serotonin transporter | Serotonin | C00003675 | 2 / 0 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00003675 | 5 / 3 |
| P04626 | Receptor tyrosine-protein kinase erbB-2 | TK tyrosine-protein kinase EGFR subfamily | C00003675 | 5 / 9 |
| P20309 | Muscarinic acetylcholine receptor M3 | Acetylcholine receptor | C00003675 | 1 / 0 |
| P21452 | Substance-K receptor | Neurokinin receptor | C00003675 | 0 / 0 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00003675 | 0 / 0 |
| P51681 | C-C chemokine receptor type 5 | CC chemokine receptor | C00003675 | 3 / 0 |
| P50406 | 5-hydroxytryptamine receptor 6 | Serotonin receptor | C00003675 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00003675 | 0 / 0 |
| P41597 | C-C chemokine receptor type 2 | CC chemokine receptor | C00003675 | 1 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00003675 | 0 / 0 |
| P08575 | Receptor-type tyrosine-protein phosphatase C | Enzyme | C00003675 | 2 / 1 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00047082 | 2 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00003675 | 3 / 3 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00003675 | 2 / 0 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00003675 | 1 / 1 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | C00003675 | 0 / 0 |
| O00204 | Sulfotransferase family cytosolic 2B member 1 | Enzyme | C00047082 | 0 / 0 |
| Q9Y4X1 | UDP-glucuronosyltransferase 2A1 | Enzyme | C00003675 | 0 / 0 |
| P06133 | UDP-glucuronosyltransferase 2B4 | Enzyme | C00003675 | 0 / 0 |
| P04062 | Glucosylceramidase | Enzyme | C00003675 | 6 / 4 |
| Q9BY64 | UDP-glucuronosyltransferase 2B28 | Enzyme | C00003675 | 0 / 0 |
| O76074 | cGMP-specific 3',5'-cyclic phosphodiesterase | PDE_5A | C00003675 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00003675 | 1 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00003675 | 1 / 0 |
| P22303 | Acetylcholinesterase | Hydrolase | C00003675 | 1 / 0 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | C00003675 | 0 / 0 |
| P28335 | 5-hydroxytryptamine receptor 2C | Serotonin receptor | C00003675 | 0 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00003675 | 0 / 0 |
| P08173 | Muscarinic acetylcholine receptor M4 | Acetylcholine receptor | C00003675 | 0 / 0 |
| P25103 | Substance-P receptor | Neurokinin receptor | C00003675 | 0 / 0 |
| P25105 | Platelet-activating factor receptor | PAF receptor | C00003675 | 0 / 0 |
| O94956 | Solute carrier organic anion transporter family member 2B1 | Unclassified protein | C00003675 | 0 / 0 |
| P33032 | Melanocortin receptor 5 | Melanocortin receptor | C00003675 | 0 / 0 |
| O15244 | Solute carrier family 22 member 2 | Drug uniporter | C00003675 | 0 / 0 |
| Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | Electrochemical transporter | C00003675 | 1 / 0 |
| P05181 | Cytochrome P450 2E1 | Cytochrome P450 2E1 | C00003675 | 0 / 0 |
| Q15125 | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | Enzyme | C00003675 | 1 / 1 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00003675 | 5 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00003675 | 4 / 3 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00003675 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00003675 | 0 / 0 |
| P10275 | Androgen receptor | NR3C4 | C00003675 | 3 / 4 |
| P23975 | Sodium-dependent noradrenaline transporter | Norepinephrine | C00003675 | 1 / 1 |
| P25100 | Alpha-1D adrenergic receptor | Adrenergic receptor | C00003675 | 0 / 0 |
| P30542 | Adenosine receptor A1 | Adenosine receptor | C00003675 | 0 / 0 |
| P18089 | Alpha-2B adrenergic receptor | Adrenergic receptor | C00003675 | 0 / 0 |
| P24557 | Thromboxane-A synthase | Cytochrome P450 5A1 | C00003675 | 1 / 1 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00003675 | 0 / 1 |
| P25025 | C-X-C chemokine receptor type 2 | CXC chemokine receptor | C00003675 | 0 / 0 |
| P49888 | Estrogen sulfotransferase | Enzyme | C00047082 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00003675 | 0 / 0 |
| P56937 | 3-keto-steroid reductase | Enzyme | C00047082 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00003675 | 1 / 1 |
| Q9NY46 | Sodium channel protein type 3 subunit alpha | SCN alpha, NaV1.x | C00003675 | 0 / 0 |
| Q99250 | Sodium channel protein type 2 subunit alpha | SCN alpha, NaV1.x | C00003675 | 2 / 2 |
| P35498 | Sodium channel protein type 1 subunit alpha | SCN alpha, NaV1.x | C00003675 | 3 / 2 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00003675 | 1 / 1 |
1243 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1583 | CYP11A1, CYP11A, CYPXIA1, P450SCC | cytochrome P450, family 11, subfamily A, polypeptide 1 (EC:1.14.15.6) |
C00003675
C00047082
|
| 1586 | CYP17A1, CPT7, CYP17, P450C17, S17AH | cytochrome P450, family 17, subfamily A, polypeptide 1 (EC:1.14.99.9 4.1.2.30) |
C00003675
C00047082
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00003675
C00047082
|
| 3479 | IGF1, IGF-I, IGF1A, IGFI | insulin-like growth factor 1 (somatomedin C) |
C00003675
C00047082
|
| 2488 | FSHB | follicle stimulating hormone, beta polypeptide |
C00003675
C00047082
|
| 3283 | HSD3B1, 3BETAHSD, HSD3B, HSDB3, HSDB3A, I, SDR11E1 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (EC:1.1.1.145 5.3.3.1) |
C00003675
C00047082
|
| 6759 | SSX4, CT5.4 | synovial sarcoma, X breakpoint 4 |
C00003675
|
| 2254 | FGF9, FGF-9, GAF, HBFG-9, HBGF-9, SYNS3 | fibroblast growth factor 9 |
C00047082
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00047082
|
| 26154 | ABCA12, ARCI4A, ARCI4B, ICR2B, LI2 | ATP-binding cassette, sub-family A (ABC1), member 12 |
C00003675
|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00003675
|
| 5825 | ABCD3, ABC43, PMP70, PXMP1, ZWS2 | ATP-binding cassette, sub-family D (ALD), member 3 |
C00003675
|
| 9429 | ABCG2, ABC15, ABCP, BCRP, BCRP1, BMDP, CD338, CDw338, EST157481, GOUT1, MRX, MXR, MXR1, UAQTL1 | ATP-binding cassette, sub-family G (WHITE), member 2 |
C00003675
|
| 140701 | ABHD16B, C20orf135, dJ591C20.1 | abhydrolase domain containing 16B |
C00003675
|
| 171586 | ABHD3, LABH3 | abhydrolase domain containing 3 |
C00003675
|
| 25 | ABL1, ABL, JTK7, bcr/abl, c-ABL, p150, v-abl | c-abl oncogene 1, non-receptor tyrosine kinase (EC:2.7.10.2) |
C00003675
|
| 80325 | ABTB1, BPOZ, BTB3, BTBD21, EF1ABP | ankyrin repeat and BTB (POZ) domain containing 1 |
C00003675
|
| 79777 | ACBD4 | acyl-CoA binding domain containing 4 |
C00003675
|
| 414149 | ACBD7, bA455B2.2 | acyl-CoA binding domain containing 7 |
C00003675
|
| 57007 | ACKR3, CMKOR1, CXC-R7, CXCR-7, CXCR7, GPR159, RDC-1, RDC1 | atypical chemokine receptor 3 |
C00003675
|
| 26027 | ACOT11, BFIT, STARD14, THEA, THEM1 | acyl-CoA thioesterase 11 (EC:3.1.2.-) |
C00003675
|
| 8309 | ACOX2, BCOX, BRCACOX, BRCOX, THCCox | acyl-CoA oxidase 2, branched chain (EC:1.17.99.3) |
C00003675
|
| 8310 | ACOX3 | acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6) |
C00003675
|
| 2181 | ACSL3, ACS3, FACL3, PRO2194 | acyl-CoA synthetase long-chain family member 3 (EC:6.2.1.3) |
C00003675
|
| 79611 | ACSS3 | acyl-CoA synthetase short-chain family member 3 (EC:6.2.1.1) |
C00003675
|
| 58 | ACTA1, ACTA, ASMA, CFTD, CFTD1, CFTDM, MPFD, NEM1, NEM2, NEM3 | actin, alpha 1, skeletal muscle |
C00003675
|
| 59 | ACTA2, AAT6, ACTSA, MYMY5 | actin, alpha 2, smooth muscle, aorta |
C00003675
|
| 72 | ACTG2, ACT, ACTA3, ACTE, ACTL3, ACTSG | actin, gamma 2, smooth muscle, enteric |
C00003675
|
| 2515 | ADAM2, CRYN1, CRYN2, CT15, FTNB, PH-30b, PH30, PH30-beta | ADAM metallopeptidase domain 2 |
C00003675
|
| 53616 | ADAM22, ADAM_22, MDC2 | ADAM metallopeptidase domain 22 |
C00003675
|
| 9370 | ADIPOQ, ACDC, ACRP30, ADIPQTL1, ADPN, APM-1, APM1, GBP28 | adiponectin, C1Q and collagen domain containing |
C00003675
|
| 134 | ADORA1, RDC7 | adenosine A1 receptor |
C00003675
|
| 136 | ADORA2B, ADORA2 | adenosine A2b receptor |
C00003675
|
| 3899 | AFF3, LAF4, MLLT2-like | AF4/FMR2 family, member 3 |
C00003675
|
| 10551 | AGR2, AG2, GOB-4, HAG-2, PDIA17, XAG-2 | anterior gradient 2 |
C00003675
|
| 79026 | AHNAK, AHNAKRS | AHNAK nucleoprotein |
C00003675
|
| 9590 | AKAP12, AKAP250, SSeCKS | A kinase (PRKA) anchor protein 12 |
C00003675
|
| 1645 | AKR1C1, 2-ALPHA-HSD, 20-ALPHA-HSD, C9, DD1, DD1/DD2, DDH, DDH1, H-37, HAKRC, HBAB, MBAB | aldo-keto reductase family 1, member C1 (EC:1.3.1.20 1.1.1.149 1.1.1.112) |
C00003675
|
| 8644 | AKR1C3, DD3, DDX, HA1753, HAKRB, HAKRe, HSD17B5, PGFS, hluPGFS | aldo-keto reductase family 1, member C3 (EC:1.3.1.20 1.1.1.188 1.1.1.239 1.1.1.64 1.1.1.112 1.1.1.357) |
C00003675
|
| 6718 | AKR1D1, 3o5bred, CBAS2, SRD5B1 | aldo-keto reductase family 1, member D1 (EC:1.3.1.3) |
C00003675
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00003675
|
| 208 | AKT2, HIHGHH, PKBB, PKBBETA, PRKBB, RAC-BETA | v-akt murine thymoma viral oncogene homolog 2 (EC:2.7.11.1) |
C00003675
|
| 213 | ALB, PRO0883, PRO0903, PRO1341 | albumin |
C00003675
|
| 220 | ALDH1A3, ALDH1A6, ALDH6, MCOP8, RALDH3 | aldehyde dehydrogenase 1 family, member A3 (EC:1.2.1.5) |
C00003675
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00003675
|
| 144245 | ALG10B, ALG10, KCR1 | ALG10B, alpha-1,2-glucosyltransferase (EC:2.4.1.256) |
C00003675
|
| 121642 | ALKBH2, ABH2 | alkB, alkylation repair homolog 2 (E. coli) (EC:1.14.11.33) |
C00003675
|
| 247 | ALOX15B, 15-LOX-2 | arachidonate 15-lipoxygenase, type B (EC:1.13.11.33) |
C00003675
|
| 115701 | ALPK2, HAK | alpha-kinase 2 |
C00003675
|
| 57538 | ALPK3, MAK, MIDORI | alpha-kinase 3 |
C00003675
|
| 250 | ALPP, ALP, PALP, PLAP, PLAP-1 | alkaline phosphatase, placental (EC:3.1.3.1) |
C00003675
|
| 259173 | ALS2CL, RN49018 | ALS2 C-terminal like |
C00003675
|
| 23600 | AMACR, AMACRD, CBAS4, RACE, RM | alpha-methylacyl-CoA racemase (EC:5.1.99.4) |
C00003675
|
| 285 | ANGPT2, AGPT2, ANG2 | angiopoietin 2 |
C00003675
|
| 286 | ANK1, ANK, SPH1, SPH2 | ankyrin 1, erythrocytic |
C00003675
|
| 56172 | ANKH, ANK, CCAL2, CMDJ, CPPDD, HANK, MANK | ANKH inorganic pyrophosphate transport regulator |
C00003675
|
| 26287 | ANKRD2, ARPP | ankyrin repeat domain 2 (stretch responsive muscle) |
C00003675
|
| 118932 | ANKRD22 | ankyrin repeat domain 22 |
C00003675
|
| 353322 | ANKRD37, Lrp2bp | ankyrin repeat domain 37 |
C00003675
|
| 54443 | ANLN, Scraps, scra | anillin, actin binding protein |
C00003675
|
| 55129 | ANO10, SCAR10, TMEM16K | anoctamin 10 |
C00003675
|
| 81611 | ANP32E, LANP-L, LANPL | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
C00003675
|
| 84168 | ANTXR1, ATR, GAPO, TEM8 | anthrax toxin receptor 1 |
C00003675
|
| 302 | ANXA2, ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV | annexin A2 |
C00003675
|
| 306 | ANXA3, ANX3 | annexin A3 (EC:3.1.4.43) |
C00003675
|
| 83464 | APH1B, APH-1B, PRO1328, PSFL, TAAV688 | APH1B gamma secretase subunit |
C00003675
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00003675
|
| 319 | APOF, Apo-F, LTIP | apolipoprotein F |
C00003675
|
| 8542 | APOL1, APO-L, APOL, APOL-I, FSGS4 | apolipoprotein L, 1 |
C00003675
|
| 10513 | APPBP2, APP-BP2, HS.84084, PAT1 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
C00003675
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00003675
|
| 116984 | ARAP2, CENTD1, PARX | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
C00003675
|
| 374 | AREG, AR, CRDGF, SDGF | amphiregulin |
C00003675
|
| 383 | ARG1 | arginase 1 (EC:3.5.3.1) |
C00003675
|
| 9824 | ARHGAP11A, GAP_(1-12) | Rho GTPase activating protein 11A |
C00003675
|
| 89839 | ARHGAP11B, B'-T, FAM7B1 | Rho GTPase activating protein 11B |
C00003675
|
| 84986 | ARHGAP19 | Rho GTPase activating protein 19 |
C00003675
|
| 23370 | ARHGEF18, P114-RhoGEF | Rho/Rac guanine nucleotide exchange factor (GEF) 18 |
C00003675
|
| 389337 | ARHGEF37 | Rho guanine nucleotide exchange factor (GEF) 37 |
C00003675
|
| 84904 | ARHGEF39, C9orf100, RP11-331F9.7 | Rho guanine nucleotide exchange factor (GEF) 39 |
C00003675
|
| 151188 | ARL6IP6, AIP-6, PFAAP1 | ADP-ribosylation-like factor 6 interacting protein 6 |
C00003675
|
| 79637 | ARMC7 | armadillo repeat containing 7 |
C00003675
|
| 10095 | ARPC1B, ARC41, p40-ARC, p41-ARC | actin related protein 2/3 complex, subunit 1B, 41kDa |
C00003675
|
| 57561 | ARRDC3, TLIMP | arrestin domain containing 3 |
C00003675
|
| 410 | ARSA, MLD | arylsulfatase A (EC:3.1.6.8) |
C00003675
|
| 55723 | ASF1B, CIA-II | anti-silencing function 1B histone chaperone |
C00003675
|
| 54529 | ASNSD1, NBLA00058, NS3TP1 | asparagine synthetase domain containing 1 |
C00003675
|
| 444 | ASPH, AAH, BAH, CASQ2BP1, HAAH, JCTN, junctin | aspartate beta-hydroxylase (EC:1.14.11.16) |
C00003675
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00003675
|
| 80150 | ASRGL1, ALP, ALP1, CRASH | asparaginase like 1 (EC:3.5.1.1 3.4.19.5) |
C00003675
|
| 79915 | ATAD5, C17orf41, ELG1, FRAG1 | ATPase family, AAA domain containing 5 |
C00003675
|
| 467 | ATF3 | activating transcription factor 3 |
C00003675
|
| 477 | ATP1A2, FHM2, MHP2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide (EC:3.6.3.9) |
C00003675
|
| 481 | ATP1B1, ATP1B | ATPase, Na+/K+ transporting, beta 1 polypeptide |
C00003675
|
| 490 | ATP2B1, PMCA1, PMCA1kb | ATPase, Ca++ transporting, plasma membrane 1 (EC:3.6.3.8) |
C00003675
|
| 10396 | ATP8A1, ATPASEII, ATPIA, ATPP2 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (EC:3.6.3.1) |
C00003675
|
| 127002 | ATXN7L2 | ataxin 7-like 2 |
C00003675
|
| 79000 | AUNIP, AIBP, C1orf135 | aurora kinase A and ninein interacting protein |
C00003675
|
| 6790 | AURKA, AIK, ARK1, AURA, AURORA2, BTAK, PPP1R47, STK15, STK6, STK7 | aurora kinase A (EC:2.7.11.1) |
C00003675
|
| 9212 | AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, PPP1R48, STK12, STK5, aurkb-sv1, aurkb-sv2 | aurora kinase B (EC:2.7.11.1) |
C00003675
|
| 8705 | B3GALT4, BETA3GALT4, GALT2, GALT4 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (EC:2.4.1.62) |
C00003675
|
| 10678 | B3GNT2, B3GN-T2, B3GNT, B3GNT-2, B3GNT1, BETA3GNT, BGNT2, BGnT-2 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (EC:2.4.1.149) |
C00003675
|
| 576 | BAI2 | brain-specific angiogenesis inhibitor 2 |
C00003675
|
| 580 | BARD1 | BRCA1 associated RING domain 1 |
C00003675
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00003675
|
| 55973 | BCAP29, BAP29 | B-cell receptor-associated protein 29 |
C00003675
|
| 8537 | BCAS1, AIBC1, NABC1 | breast carcinoma amplified sequence 1 |
C00003675
|
| 590 | BCHE, CHE1, CHE2, E1 | butyrylcholinesterase (EC:3.1.1.8) |
C00003675
|
| 8915 | BCL10, CARMEN, CIPER, CLAP, c-E10, mE10 | B-cell CLL/lymphoma 10 |
C00003675
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00003675
|
| 83596 | BCL2L12 | BCL2-like 12 (proline rich) |
C00003675
|
| 623 | BDKRB1, B1BKR, B1R, BKB1R, BKR1, BRADYB1 | bradykinin receptor B1 |
C00003675
|
| 330 | BIRC3, AIP1, API2, CIAP2, HAIP1, HIAP1, MALT2, MIHC, RNF49, c-IAP2 | baculoviral IAP repeat containing 3 |
C00003675
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00003675
|
| 641 | BLM, BS, RECQ2, RECQL2, RECQL3 | Bloom syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00003675
|
| 658 | BMPR1B, ALK-6, ALK6, CDw293 | bone morphogenetic protein receptor, type IB (EC:2.7.11.30) |
C00003675
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00003675
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00003675
|
| 83990 | BRIP1, BACH1, FANCJ, OF | BRCA1 interacting protein C-terminal helicase 1 (EC:3.6.4.13) |
C00003675
|
| 55299 | BRIX1, BRIX, BXDC2 | BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
C00003675
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00003675
|
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00003675
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00003675
|
| 11067 | C10orf10, DEPP, FIG | chromosome 10 open reading frame 10 |
C00003675
|
| 79703 | C11orf80 | chromosome 11 open reading frame 80 |
C00003675
|
| 220042 | C11orf82, noxin | chromosome 11 open reading frame 82 |
C00003675
|
| 121053 | C12orf45 | chromosome 12 open reading frame 45 |
C00003675
|
| 79794 | C12orf49 | chromosome 12 open reading frame 49 |
C00003675
|
| 730094 | C16orf52 | chromosome 16 open reading frame 52 |
C00003675
|
| 80178 | C16orf59 | chromosome 16 open reading frame 59 |
C00003675
|
| 78995 | C17orf53 | chromosome 17 open reading frame 53 |
C00003675
|
| 162681 | C18orf54, LAS2 | chromosome 18 open reading frame 54 |
C00003675
|
| 494514 | C18orf56 | chromosome 18 open reading frame 56 |
C00003675
|
| 55732 | C1orf112, RP1-97P20.1 | chromosome 1 open reading frame 112 |
C00003675
|
| 81563 | C1orf21, PIG13 | chromosome 1 open reading frame 21 |
C00003675
|
| 140688 | C20orf112, C20orf113, dJ1184F4.2, dJ1184F4.4 | chromosome 20 open reading frame 112 |
C00003675
|
| 54976 | C20orf27 | chromosome 20 open reading frame 27 |
C00003675
|
| 54058 | C21orf58 | chromosome 21 open reading frame 58 |
C00003675
|
| 55345 | C4orf21 | chromosome 4 open reading frame 21 |
C00003675
|
| 201725 | C4orf46 | chromosome 4 open reading frame 46 |
C00003675
|
| 375444 | C5orf34 | chromosome 5 open reading frame 34 |
C00003675
|
| 51149 | C5orf45 | chromosome 5 open reading frame 45 |
C00003675
|
| 79846 | C7orf63 | chromosome 7 open reading frame 63 |
C00003675
|
| 56892 | C8orf4, TC-1, TC1 | chromosome 8 open reading frame 4 |
C00003675
|
| 55071 | C9orf40 | chromosome 9 open reading frame 40 |
C00003675
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00003675
|
| 81617 | CAB39L, MO25-BETA, MO2L, bA103J18.3 | calcium binding protein 39-like |
C00003675
|
| 27092 | CACNG4 | calcium channel, voltage-dependent, gamma subunit 4 |
C00003675
|
| 93664 | CADPS2 | Ca++-dependent secretion activator 2 |
C00003675
|
| 811 | CALR, CRT, RO, SSA, cC1qR | calreticulin |
C00003675
|
| 57118 | CAMK1D, CKLiK, CaM-K1, CaMKID | calcium/calmodulin-dependent protein kinase ID (EC:2.7.11.17) |
C00003675
|
| 23261 | CAMTA1, CANPMR | calmodulin binding transcription activator 1 |
C00003675
|
| 147968 | CAPN12 | calpain 12 |
C00003675
|
| 824 | CAPN2, CANP2, CANPL2, CANPml, mCANP | calpain 2, (m/II) large subunit (EC:3.4.22.53) |
C00003675
|
| 84674 | CARD6, CINCIN1 | caspase recruitment domain family, member 6 |
C00003675
|
| 57082 | CASC5, AF15Q14, CT29, D40, KNL1, PPP1R55, Spc7, hKNL-1, hSpc105 | cancer susceptibility candidate 5 |
C00003675
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00003675
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00003675
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00003675
|
| 857 | CAV1, BSCL3, CGL3, MSTP085, PPH3, VIP21, CAV | caveolin 1, caveolae protein, 22kDa |
C00003675
|
| 858 | CAV2, CAV | caveolin 2 |
C00003675
|
| 79872 | CBLL1, HAKAI, RNF188 | Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
C00003675
|
| 147381 | CBLN2 | cerebellin 2 precursor |
C00003675
|
| 84733 | CBX2, CDCA6, M33, SRXY5 | chromobox homolog 2 |
C00003675
|
| 165055 | CCDC138 | coiled-coil domain containing 138 |
C00003675
|
| 64770 | CCDC14 | coiled-coil domain containing 14 |
C00003675
|
| 80071 | CCDC15 | coiled-coil domain containing 15 |
C00003675
|
| 343099 | CCDC18, NY-SAR-41, dJ717I23.1 | coiled-coil domain containing 18 |
C00003675
|
| 149473 | CCDC24, RP5-1198O20.2 | coiled-coil domain containing 24 |
C00003675
|
| 91057 | CCDC34, NY-REN-41, RAMA3 | coiled-coil domain containing 34 |
C00003675
|
| 92558 | CCDC64, BICDR-1, H_267D11.1 | coiled-coil domain containing 64 |
C00003675
|
| 114800 | CCDC85A | coiled-coil domain containing 85A |
C00003675
|
| 6355 | CCL8, HC14, MCP-2, MCP2, SCYA10, SCYA8 | chemokine (C-C motif) ligand 8 |
C00003675
|
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00003675
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00003675
|
| 9133 | CCNB2, HsT17299 | cyclin B2 |
C00003675
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00003675
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00003675
|
| 126731 | CCSAP, C1orf96, CSAP | centriole, cilia and spindle-associated protein |
C00003675
|
| 929 | CD14 | CD14 molecule |
C00003675
|
| 100133941 | CD24, CD24A | CD24 molecule |
C00003675
|
| 29126 | CD274, B7-H, B7H1, PD-L1, PDCD1L1, PDCD1LG1, PDL1 | CD274 molecule |
C00003675
|
| 966 | CD59, 16.3A5, 1F5, EJ16, EJ30, EL32, G344, HRF-20, HRF20, MAC-IP, MACIF, MEM43, MIC11, MIN1, MIN2, MIN3, MIRL, MSK21, p18-20 | CD59 molecule, complement regulatory protein |
C00003675
|
| 968 | CD68, GP110, LAMP4, SCARD1 | CD68 molecule |
C00003675
|
| 924 | CD7, GP40, LEU-9, TP41, Tp40 | CD7 molecule |
C00003675
|
| 3732 | CD82, 4F9, C33, GR15, IA4, KAI1, R2, SAR2, ST6, TSPAN27 | CD82 molecule |
C00003675
|
| 34411 |
C00003675
|
||
| 991 | CDC20, CDC20A, bA276H19.3, p55CDC | cell division cycle 20 |
C00003675
|
| 994 | CDC25B | cell division cycle 25B (EC:3.1.3.48) |
C00003675
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00003675
|
| 10602 | CDC42EP3, BORG2, CEP3, UB1 | CDC42 effector protein (Rho GTPase binding) 3 |
C00003675
|
| 31052 |
C00003675
|
||
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00003675
|
| 157313 | CDCA2, Repo-Man | cell division cycle associated 2 |
C00003675
|
| 83461 | CDCA3, GRCC8, TOME-1 | cell division cycle associated 3 |
C00003675
|
| 55038 | CDCA4, HEPP, SEI-3/HEPP | cell division cycle associated 4 |
C00003675
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00003675
|
| 83879 | CDCA7, JPO1 | cell division cycle associated 7 |
C00003675
|
| 55536 | CDCA7L, JPO2, R1, RAM2 | cell division cycle associated 7-like |
C00003675
|
| 55143 | CDCA8, BOR, BOREALIN, DasraB, MESRGP | cell division cycle associated 8 |
C00003675
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00003675
|
| 1030 | CDKN2B, CDK4I, INK4B, MTS2, P15, TP15, p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
C00003675
|
| 1031 | CDKN2C, INK4C, p18, p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
C00003675
|
| 1032 | CDKN2D, INK4D, p19, p19-INK4D | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
C00003675
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00003675
|
| 284040 | CDRT4 | CMT1A duplicated region transcript 4 |
C00003675
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00003675
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00003675
|
| 27439 | CECR6 | cat eye syndrome chromosome region, candidate 6 |
C00003675
|
| 1056 | CEL, BAL, BSDL, BSSL, CELL, CEase, FAP, FAPP, LIPA, MODY8 | carboxyl ester lipase (EC:3.1.1.13 3.1.1.3) |
C00003675
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00003675
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00003675
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00003675
|
| 64946 | CENPH | centromere protein H |
C00003675
|
| 2491 | CENPI, CENP-I, FSHPRH1, LRPR1, Mis6 | centromere protein I |
C00003675
|
| 55835 | CENPJ, BM032, CENP-J, CPAP, LAP, LIP1, MCPH6, SASS4, SCKL4, Sas-4 | centromere protein J |
C00003675
|
| 64105 | CENPK, AF5alpha, CENP-K, P33, Solt | centromere protein K |
C00003675
|
| 91687 | CENPL, C1orf155, CENP-L, RP3-383J4.1, dJ383J4.3 | centromere protein L |
C00003675
|
| 79019 | CENPM, C22orf18, CENP-M, PANE1, bK250D10.2 | centromere protein M |
C00003675
|
| 79172 | CENPO, CENP-O, ICEN-36, MCM21R | centromere protein O |
C00003675
|
| 55166 | CENPQ, C6orf139, CENP-Q | centromere protein Q |
C00003675
|
| 387103 | CENPW, C6orf173, CENP-W, CUG2 | centromere protein W |
C00003675
|
| 145508 | CEP128, C14orf145, C14orf61, LEDP/132 | centrosomal protein 128kDa |
C00003675
|
| 22995 | CEP152, MCPH4, MCPH9, SCKL5 | centrosomal protein 152kDa |
C00003675
|
| 9857 | CEP350, CAP350, GM133 | centrosomal protein 350kDa |
C00003675
|
| 55165 | CEP55, C10orf3, CT111, URCC6 | centrosomal protein 55kDa |
C00003675
|
| 285753 | CEP57L1, C6orf182, bA487F23.2, cep57R | centrosomal protein 57kDa-like 1 |
C00003675
|
| 80254 | CEP63 | centrosomal protein 63kDa |
C00003675
|
| 55722 | CEP72 | centrosomal protein 72kDa |
C00003675
|
| 84131 | CEP78, C9orf81, IP63 | centrosomal protein 78kDa |
C00003675
|
| 1081 | CGA, CG-ALPHA, FSHA, GPHA1, GPHa, HCG, LHA, TSHA | glycoprotein hormones, alpha polypeptide |
C00003675
|
| 1082 | CGB, CGB3, CGB5, CGB7, CGB8, hCGB | chorionic gonadotropin, beta polypeptide |
C00003675
|
| 494143 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) |
C00003675
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00003675
|
| 8208 | CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7, MPP7 | chromatin assembly factor 1, subunit B (p60) |
C00003675
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00003675
|
| 79586 | CHPF, CHSY2, CSS2 | chondroitin polymerizing factor (EC:2.4.1.175 2.4.1.226) |
C00003675
|
| 1131 | CHRM3, EGBRS, HM3 | cholinergic receptor, muscarinic 3 |
C00003675
|
| 1135 | CHRNA2 | cholinergic receptor, nicotinic, alpha 2 (neuronal) |
C00003675
|
| 1145 | CHRNE, ACHRE, CMS1D, CMS1E, CMS2A, FCCMS, SCCMS | cholinergic receptor, nicotinic, epsilon (muscle) |
C00003675
|
| 63922 | CHTF18, C16orf41, C321D2.2, C321D2.3, C321D2.4, CHL12, Ctf18, RUVBL | CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
C00003675
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00003675
|
| 26586 | CKAP2, LB1, TMAP, se20-10 | cytoskeleton associated protein 2 |
C00003675
|
| 150468 | CKAP2L | cytoskeleton associated protein 2-like |
C00003675
|
| 51192 | CKLF, C32, CKLF1, CKLF2, CKLF3, CKLF4, UCK-1 | chemokine-like factor |
C00003675
|
| 1163 | CKS1B, CKS1, PNAS-16, PNAS-18, ckshs1 | CDC28 protein kinase regulatory subunit 1B |
C00003675
|
| 1164 | CKS2, CKSHS2 | CDC28 protein kinase regulatory subunit 2 |
C00003675
|
| 1180 | CLCN1, CLC1 | chloride channel, voltage-sensitive 1 |
C00003675
|
| 9069 | CLDN12 | claudin 12 |
C00003675
|
| 1364 | CLDN4, CPE-R, CPER, CPETR, CPETR1, WBSCR8, hCPE-R | claudin 4 |
C00003675
|
| 9073 | CLDN8 | claudin 8 |
C00003675
|
| 7123 | CLEC3B, TN, TNA | C-type lectin domain family 3, member B |
C00003675
|
| 79745 | CLIP4, RSNL2 | CAP-GLY domain containing linker protein family, member 4 |
C00003675
|
| 79789 | CLMN | calmin (calponin-like, transmembrane) |
C00003675
|
| 63967 | CLSPN | claspin |
C00003675
|
| 134147 | CMBL, JS-1 | carboxymethylenebutenolidase homolog (Pseudomonas) (EC:3.1.-.-) |
C00003675
|
| 84319 | CMSS1, C3orf26 | cms1 ribosomal small subunit homolog (yeast) |
C00003675
|
| 254263 | CNIH2, CNIH-2, Cnil | cornichon family AMPA receptor auxiliary protein 2 |
C00003675
|
| 1265 | CNN2 | calponin 2 |
C00003675
|
| 1281 | COL3A1, EDS4A | collagen, type III, alpha 1 |
C00003675
|
| 120376 | COLCA2, C11orf93, CASC13 | colorectal cancer associated 2 |
C00003675
|
| 81035 | COLEC12, CLP1, NSR2, SCARA4, SRCL | collectin sub-family member 12 |
C00003675
|
| 55352 | COPRS, C17orf79, COPR5, HSA272196, TTP1 | coordinator of PRMT5, differentiation stimulator |
C00003675
|
| 51805 | COQ3, DHHBMT, DHHBMTASE, UG0215E05, bA9819.1 | coenzyme Q3 methyltransferase (EC:2.1.1.114 2.1.1.64 2.1.1.222) |
C00003675
|
| 170712 | COX7B2 | cytochrome c oxidase subunit VIIb2 (EC:1.9.3.1) |
C00003675
|
| 33136 |
C00003675
|
||
| 1362 | CPD, GP180 | carboxypeptidase D (EC:3.4.17.22) |
C00003675
|
| 132864 | CPEB2, CPE-BP2, CPEB-2, hCPEB-2 | cytoplasmic polyadenylation element binding protein 2 |
C00003675
|
| 22849 | CPEB3 | cytoplasmic polyadenylation element binding protein 3 |
C00003675
|
| 10488 | CREB3, LUMAN, LZIP | cAMP responsive element binding protein 3 |
C00003675
|
| 90993 | CREB3L1, OASIS | cAMP responsive element binding protein 3-like 1 |
C00003675
|
| 1387 | CREBBP, CBP, KAT3A, RSTS | CREB binding protein (EC:2.3.1.48) |
C00003675
|
| 1410 | CRYAB, CMD1II, CRYA2, CTPP2, CTRCT16, HSPB5, MFM2 | crystallin, alpha B |
C00003675
|
| 1434 | CSE1L, CAS, CSE1, XPO2 | CSE1 chromosome segregation 1-like (yeast) |
C00003675
|
| 1435 | CSF1, CSF-1, MCSF | colony stimulating factor 1 (macrophage) (EC:2.7.10.1) |
C00003675
|
| 55790 | CSGALNACT1, CSGalNAcT-1, ChGn, beta4GalNAcT | chondroitin sulfate N-acetylgalactosaminyltransferase 1 (EC:2.4.1.174) |
C00003675
|
| 64651 | CSRNP1, AXUD1, CSRNP-1, FAM130B, TAIP-3, URAX1 | cysteine-serine-rich nuclear protein 1 |
C00003675
|
| 1496 | CTNNA2, CAP-R, CAPR, CT114, CTNR | catenin (cadherin-associated protein), alpha 2 |
C00003675
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00003675
|
| 1515 | CTSV, CATL2, CTSL2, CTSU | cathepsin V (EC:3.4.22.43) |
C00003675
|
| 348180 | CTU2, C16orf84, NCS2, UPF0432 | cytosolic thiouridylase subunit 2 homolog (S. pombe) |
C00003675
|
| 1525 | CXADR, CAR, CAR4/6, HCAR | coxsackie virus and adenovirus receptor |
C00003675
|
| 6373 | CXCL11, H174, I-TAC, IP-9, IP9, SCYB11, SCYB9B, b-R1 | chemokine (C-X-C motif) ligand 11 |
C00003675
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00003675
|
| 284613 | CYB561D1 | cytochrome b561 family, member D1 |
C00003675
|
| 11068 | CYB561D2, 101F6, TSP10 | cytochrome b561 family, member D2 |
C00003675
|
| 1585 | CYP11B2, ALDOS, CPN2, CYP11B, CYP11BL, CYPXIB2, P-450C18, P450C18, P450aldo | cytochrome P450, family 11, subfamily B, polypeptide 2 (EC:1.14.15.4 1.14.15.5) |
C00003675
|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00003675
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00003675
|
| 1591 | CYP24A1, CP24, CYP24, HCAI, P450-CC24 | cytochrome P450, family 24, subfamily A, polypeptide 1 (EC:1.14.13.126) |
C00003675
|
| 1548 | CYP2A6, CPA6, CYP2A, CYP2A3, CYPIIA6, P450C2A, P450PB | cytochrome P450, family 2, subfamily A, polypeptide 6 (EC:1.14.14.1) |
C00003675
|
| 1549 | CYP2A7, CPA7, CPAD, CYP2A, CYPIIA7, P450-IIA4 | cytochrome P450, family 2, subfamily A, polypeptide 7 (EC:1.14.14.1) |
C00003675
|
| 1555 | CYP2B6, CPB6, CYP2B, CYP2B7, CYP2B7P, CYPIIB6, EFVM, IIB1, P450 | cytochrome P450, family 2, subfamily B, polypeptide 6 (EC:1.14.14.1) |
C00003675
|
| 29785 | CYP2S1 | cytochrome P450, family 2, subfamily S, polypeptide 1 (EC:1.14.14.1) |
C00003675
|
| 113612 | CYP2U1, P450TEC, SPG49, SPG56 | cytochrome P450, family 2, subfamily U, polypeptide 1 (EC:1.14.14.1) |
C00003675
|
| 1574 |
C00003675
|
||
| 64816 | CYP3A43 | cytochrome P450, family 3, subfamily A, polypeptide 43 (EC:1.14.14.1) |
C00003675
|
| 1577 | CYP3A5, CP35, CYPIIIA5, P450PCN3, PCN3 | cytochrome P450, family 3, subfamily A, polypeptide 5 (EC:1.14.14.1) |
C00003675
|
| 1551 | CYP3A7, CP37, CYPIIIA7, P450-HFLA | cytochrome P450, family 3, subfamily A, polypeptide 7 (EC:1.14.14.1) |
C00003675
|
| 10858 | CYP46A1, CP46, CYP46 | cytochrome P450, family 46, subfamily A, polypeptide 1 (EC:1.14.13.98) |
C00003675
|
| 84418 | CYSTM1, C5orf32 | cysteine-rich transmembrane module containing 1 |
C00003675
|
| 1612 | DAPK1, DAPK | death-associated protein kinase 1 (EC:2.7.11.1) |
C00003675
|
| 23604 | DAPK2, DRP-1, DRP1 | death-associated protein kinase 2 (EC:2.7.11.1) |
C00003675
|
| 10926 | DBF4, ASK, CHIF, DBF4A, ZDBF1 | DBF4 homolog (S. cerevisiae) |
C00003675
|
| 80174 | DBF4B, ASKL1, CHIFB, DRF1, ZDBF1B | DBF4 homolog B (S. cerevisiae) |
C00003675
|
| 100528464 |
C00003675
|
||
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00003675
|
| 1663 | DDX11, CHL1, CHLR1, KRG2, WABS | DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 (EC:3.6.4.13) |
C00003675
|
| 68278 |
C00003675
|
||
| 7913 | DEK, D6S231E | DEK oncogene |
C00003675
|
| 79958 | DENND1C, FAM31C | DENN/MADD domain containing 1C |
C00003675
|
| 55635 | DEPDC1, DEP.8, DEPDC1-V2, DEPDC1A, SDP35 | DEP domain containing 1 |
C00003675
|
| 55789 | DEPDC1B, BRCC3, XTP1 | DEP domain containing 1B |
C00003675
|
| 1677 | DFFB, CAD, CPAN, DFF-40, DFF2, DFF40 | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
C00003675
|
| 1606 | DGKA, DAGK, DAGK1, DGK-alpha | diacylglycerol kinase, alpha 80kDa (EC:2.7.1.107) |
C00003675
|
| 160851 | DGKH, DGKeta | diacylglycerol kinase, eta (EC:2.7.1.107) |
C00003675
|
| 10202 | DHRS2, HEP27, SDR25C1 | dehydrogenase/reductase (SDR family) member 2 |
C00003675
|
| 9249 | DHRS3, DD83.1, RDH17, Rsdr1, SDR1, SDR16C1, retSDR1 | dehydrogenase/reductase (SDR family) member 3 (EC:1.1.1.300) |
C00003675
|
| 81624 | DIAPH3, AN, AUNA1, DIA2, DRF3, NSDAN, diap3, mDia2 | diaphanous-related formin 3 |
C00003675
|
| 129563 | DIS3L2, FAM6A, PRLMNS, hDIS3L2 | DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
C00003675
|
| 84976 | DISP1, DISPA | dispatched homolog 1 (Drosophila) |
C00003675
|
| 9787 | DLGAP5, DLG7, HURP | discs, large (Drosophila) homolog-associated protein 5 |
C00003675
|
| 127343 | DMBX1, MBX, OTX3, PAXB | diencephalon/mesencephalon homeobox 1 |
C00003675
|
| 29958 | DMGDH, DMGDHD, ME2GLYDH | dimethylglycine dehydrogenase (EC:1.5.8.4) |
C00003675
|
| 1763 | DNA2, DNA2L, PEOA6, hDNA2 | DNA replication helicase/nuclease 2 (EC:3.6.4.12) |
C00003675
|
| 79982 | DNAJB14, EGNR9427, PRO34683 | DnaJ (Hsp40) homolog, subfamily B, member 14 |
C00003675
|
| 414061 | DNAJB3, HCG3 | DnaJ (Hsp40) homolog, subfamily B, member 3 |
C00003675
|
| 5611 | DNAJC3, ERdj6, HP58, P58, P58IPK, PRKRI | DnaJ (Hsp40) homolog, subfamily C, member 3 |
C00003675
|
| 23234 | DNAJC9, HDJC9, JDD1, SB73 | DnaJ (Hsp40) homolog, subfamily C, member 9 |
C00003675
|
| 373863 | DND1, RBMS4 | DND microRNA-mediated repression inhibitor 1 |
C00003675
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00003675
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00003675
|
| 81704 | DOCK8, MRD2, ZIR8 | dedicator of cytokinesis 8 |
C00003675
|
| 55715 | DOK4 | docking protein 4 |
C00003675
|
| 29980 | DONSON, B17, C21orf60 | downstream neighbor of SON |
C00003675
|
| 8193 | DPF1, BAF45b, NEUD4, neuro-d4 | D4, zinc and double PHD fingers family 1 |
C00003675
|
| 1823 | DSC1, CDHF1, DG2/DG3 | desmocollin 1 |
C00003675
|
| 1824 | DSC2, ARVD11, CDHF2, DG2, DGII/III, DSC3 | desmocollin 2 |
C00003675
|
| 92126 | DSEL, C18orf4 | dermatan sulfate epimerase-like |
C00003675
|
| 51514 | DTL, CDT2, DCAF2, L2DTL, RAMP | denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
C00003675
|
| 1841 | DTYMK, CDC8, PP3731, TMPK, TYMK | deoxythymidylate kinase (thymidylate kinase) (EC:2.7.4.9) |
C00003675
|
| 53905 | DUOX1, LNOX1, NOXEF1, THOX1 | dual oxidase 1 (EC:1.6.3.1) |
C00003675
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00003675
|
| 1847 | DUSP5, DUSP, HVH3 | dual specificity phosphatase 5 (EC:3.1.3.16 3.1.3.48) |
C00003675
|
| 1849 | DUSP7, MKPX, PYST2 | dual specificity phosphatase 7 (EC:3.1.3.16 3.1.3.48) |
C00003675
|
| 1854 | DUT, dUTPase | deoxyuridine triphosphatase (EC:3.6.1.23) |
C00003675
|
| 643001 |
C00003675
|
||
| 554045 | DUX4L9, DUX4C | double homeobox 4 like 9 |
C00003675
|
| 8291 | DYSF, FER1L1, LGMD2B, MMD1 | dysferlin |
C00003675
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00003675
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00003675
|
| 144455 | E2F7 | E2F transcription factor 7 |
C00003675
|
| 79733 | E2F8, E2F-8 | E2F transcription factor 8 |
C00003675
|
| 55840 | EAF2, BM040, TRAITS, U19 | ELL associated factor 2 |
C00003675
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00003675
|
| 1906 | EDN1, ET1, HDLCQ7, PPET1 | endothelin 1 |
C00003675
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00003675
|
| 79631 | EFTUD1, FAM42A, HsT19294, RIA1 | elongation factor Tu GTP binding domain containing 1 |
C00003675
|
| 1950 | EGF, HOMG4, URG | epidermal growth factor |
C00003675
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00003675
|
| 112398 | EGLN2, EIT6, HIF-PH1, HIFPH1, HPH-1, HPH-3, PHD1 | egl-9 family hypoxia-inducible factor 2 (EC:1.14.11.29) |
C00003675
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00003675
|
| 319118 |
C00003675
|
||
| 1978 | EIF4EBP1, 4E-BP1, 4EBP1, BP-1, PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 |
C00003675
|
| 1997 | ELF1 | E74-like factor 1 (ets domain transcription factor) |
C00003675
|
| 1999 | ELF3, EPR-1, ERT, ESE-1, ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
C00003675
|
| 63916 | ELMO2, CED-12, CED12, ELMO-2 | engulfment and cell motility 2 |
C00003675
|
| 146956 | EME1, MMS4L, SLX2A | essential meiotic structure-specific endonuclease 1 |
C00003675
|
| 2012 | EMP1, CL-20, EMP-1, TMP | epithelial membrane protein 1 |
C00003675
|
| 5168 | ENPP2, ATX, ATX-X, AUTOTAXIN, LysoPLD, NPP2, PD-IALPHA, PDNP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 (EC:3.1.4.39) |
C00003675
|
| 377841 | ENTPD8, GLSR2492, NTPDase-8, UNQ2492 | ectonucleoside triphosphate diphosphohydrolase 8 (EC:3.6.1.5) |
C00003675
|
| 23136 | EPB41L3, 4.1B, DAL-1, DAL1 | erythrocyte membrane protein band 4.1-like 3 |
C00003675
|
| 2041 | EPHA1, EPH, EPHT, EPHT1 | EPH receptor A1 (EC:2.7.10.1) |
C00003675
|
| 2051 | EPHB6, HEP | EPH receptor B6 (EC:2.7.10.1) |
C00003675
|
| 2056 | EPO, EP, MVCD2 | erythropoietin |
C00003675
|
| 54869 | EPS8L1, DRC3, EPS8R1 | EPS8-like 1 |
C00003675
|
| 54821 | ERCC6L, PICH, RAD26L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like (EC:3.6.4.12) |
C00003675
|
| 30001 | ERO1L, ERO1-alpha, ERO1A, ERO1LA | ERO1-like (S. cerevisiae) |
C00003675
|
| 54206 | ERRFI1, GENE-33, MIG-6, MIG6, RALT | ERBB receptor feedback inhibitor 1 |
C00003675
|
| 157570 | ESCO2, 2410004I17Rik, EFO2, RBS | establishment of sister chromatid cohesion N-acetyltransferase 2 (EC:2.3.1.-) |
C00003675
|
| 9700 | ESPL1, ESP1, SEPA | extra spindle pole bodies homolog 1 (S. cerevisiae) (EC:3.4.22.49) |
C00003675
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00003675
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00003675
|
| 55194 | EVA1B, C1orf78, FAM176B, RP11-268J15.2 | eva-1 homolog B (C. elegans) |
C00003675
|
| 2121 | EVC, DWF-1, EVC1, EVCL | Ellis van Creveld syndrome |
C00003675
|
| 9156 | EXO1, HEX1, hExoI | exonuclease 1 |
C00003675
|
| 64789 | EXO5, C1orf176, DEM1, Exo_V, hExo5 | exonuclease 5 |
C00003675
|
| 23404 | EXOSC2, RRP4, Rrp4p, hRrp4p, p7 | exosome component 2 |
C00003675
|
| 11340 | EXOSC8, CIP3, EAP2, OIP2, RP11-421P11.3, RRP43, Rrp43p, bA421P11.3, p9 | exosome component 8 (EC:3.1.13.-) |
C00003675
|
| 5393 | EXOSC9, PM/Scl-75, PMSCL1, RRP45, Rrp45p, p5, p6 | exosome component 9 (EC:3.1.13.-) |
C00003675
|
| 2146 | EZH2, ENX-1, ENX1, EZH1, EZH2b, KMT6, KMT6A, WVS, WVS2 | enhancer of zeste homolog 2 (Drosophila) (EC:2.1.1.43) |
C00003675
|
| 2153 | F5, FVL, PCCF, RPRGL1, THPH2 | coagulation factor V (proaccelerin, labile factor) |
C00003675
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00003675
|
| 9415 | FADS2, D6D, DES6, FADSD6, LLCDL2, SLL0262, TU13 | fatty acid desaturase 2 (EC:1.14.19.-) |
C00003675
|
| 359845 | FAM101B | family with sequence similarity 101, member B |
C00003675
|
| 54491 | FAM105A, NET20 | family with sequence similarity 105, member A |
C00003675
|
| 642273 | FAM110C | family with sequence similarity 110, member C |
C00003675
|
| 374393 | FAM111B, CANP | family with sequence similarity 111, member B |
C00003675
|
| 9715 | FAM131B | family with sequence similarity 131, member B |
C00003675
|
| 9413 | FAM189A2, C9orf61, RP11-548B3.1, X123 | family with sequence similarity 189, member A2 |
C00003675
|
| 51313 | FAM198B, AD036, C4orf18 | family with sequence similarity 198, member B |
C00003675
|
| 388886 | FAM211B, C22orf36 | family with sequence similarity 211, member B |
C00003675
|
| 80256 | FAM214B, KIAA1539, P1.11659_5, RP11-182N22.6, bA182N22.6 | family with sequence similarity 214, member B |
C00003675
|
| 29902 | FAM216A, C12orf24, HSU79274 | family with sequence similarity 216, member A |
C00003675
|
| 366403 |
C00003675
|
||
| 54478 | FAM64A, CATS, RCS1 | family with sequence similarity 64, member A |
C00003675
|
| 728833 | FAM72D, GCUD2 | family with sequence similarity 72, member D |
C00003675
|
| 145773 | FAM81A | family with sequence similarity 81, member A |
C00003675
|
| 81610 | FAM83D, C20orf129, CHICA, dJ616B8.3 | family with sequence similarity 83, member D |
C00003675
|
| 54854 | FAM83E | family with sequence similarity 83, member E |
C00003675
|
| 2175 | FANCA, FA, FA-H, FA1, FAA, FACA, FAH, FANCH | Fanconi anemia, complementation group A |
C00003675
|
| 2187 | FANCB, FA2, FAAP90, FAAP95, FAB, FACB | Fanconi anemia, complementation group B |
C00003675
|
| 2176 | FANCC, FA3, FAC, FACC | Fanconi anemia, complementation group C |
C00003675
|
| 2177 | FANCD2, FA-D2, FA4, FACD, FAD, FAD2, FANCD | Fanconi anemia, complementation group D2 |
C00003675
|
| 2178 | FANCE, FACE, FAE | Fanconi anemia, complementation group E |
C00003675
|
| 2189 | FANCG, FAG, XRCC9 | Fanconi anemia, complementation group G |
C00003675
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00003675
|
| 55711 | FAR2, MLSTD1, SDR10E2 | fatty acyl CoA reductase 2 (EC:1.2.1.n2) |
C00003675
|
| 356 | FASLG, ALPS1B, APT1LG1, APTL, CD178, CD95-L, CD95L, FASL, TNFSF6 | Fas ligand (TNF superfamily, member 6) |
C00003675
|
| 54751 | FBLIM1, CAL, FBLP-1, FBLP1 | filamin binding LIM protein 1 |
C00003675
|
| 26272 | FBXO4, FBX4 | F-box protein 4 |
C00003675
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00003675
|
| 2237 | FEN1, FEN-1, MF1, RAD2 | flap structure-specific endonuclease 1 |
C00003675
|
| 9637 | FEZ2, HUM3CL | fasciculation and elongation protein zeta 2 (zygin II) |
C00003675
|
| 121512 | FGD4, CMT4H, FRABP, ZFYVE6 | FYVE, RhoGEF and PH domain containing 4 |
C00003675
|
| 2263 | FGFR2, BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 | fibroblast growth factor receptor 2 (EC:2.7.10.1) |
C00003675
|
| 2267 | FGL1, HFREP1, HP-041, LFIRE-1, LFIRE1 | fibrinogen-like 1 |
C00003675
|
| 2274 | FHL2, AAG11, DRAL, FHL-2, SLIM-3, SLIM3 | four and a half LIM domains 2 |
C00003675
|
| 63979 | FIGNL1 | fidgetin-like 1 |
C00003675
|
| 2281 | FKBP1B, FKBP12.6, FKBP1L, OTK4, PKBP1L, PPIase | FK506 binding protein 1B, 12.6 kDa (EC:5.2.1.8) |
C00003675
|
| 2289 | FKBP5, AIG6, FKBP51, FKBP54, P54, PPIase, Ptg-10 | FK506 binding protein 5 (EC:5.2.1.8) |
C00003675
|
| 2329 | FMO4, FMO2 | flavin containing monooxygenase 4 (EC:1.14.13.8) |
C00003675
|
| 2330 | FMO5 | flavin containing monooxygenase 5 (EC:1.14.13.8) |
C00003675
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00003675
|
| 2296 | FOXC1, ARA, FKHL7, FREAC-3, FREAC3, IGDA, IHG1, IRID1, RIEG3 | forkhead box C1 |
C00003675
|
| 2298 | FOXD4, FKHL9, FOXD4A, FREAC-5, FREAC5 | forkhead box D4 |
C00003675
|
| 286380 | FOXD4L3, FOXD4L2, FOXD6 | forkhead box D4-like 3 |
C00003675
|
| 2305 | FOXM1, FKHL16, FOXM1B, HFH-11, HFH11, HNF-3, INS-1, MPHOSPH2, MPP-2, MPP2, PIG29, TGT3, TRIDENT | forkhead box M1 |
C00003675
|
| 4303 | FOXO4, AFX, AFX1, MLLT7 | forkhead box O4 |
C00003675
|
| 2357 | FPR1, FMLP, FPR | formyl peptide receptor 1 |
C00003675
|
| 158326 | FREM1, BNAR, C9orf143, C9orf145, C9orf154, MOTA, TILRR, TRIGNO2 | FRAS1 related extracellular matrix 1 |
C00003675
|
| 143162 | FRMPD2, PDZD5C, PDZK4, PDZK5C | FERM and PDZ domain containing 2 |
C00003675
|
| 2521 | FUS, ALS6, ETM4, FUS1, HNRNPP2, POMP75, TLS | fused in sarcoma |
C00003675
|
| 2525 | FUT3, CD174, FT3B, FucT-III, LE, Les | fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (EC:2.4.1.65) |
C00003675
|
| 5349 | FXYD3, MAT8, PLML | FXYD domain containing ion transport regulator 3 |
C00003675
|
| 7855 | FZD5, C2orf31, HFZ5 | frizzled family receptor 5 |
C00003675
|
| 55632 | G2E3, KIAA1333, PHF7B | G2/M-phase specific E3 ubiquitin protein ligase |
C00003675
|
| 2539 | G6PD, G6PD1 | glucose-6-phosphate dehydrogenase (EC:1.1.1.49) |
C00003675
|
| 4616 | GADD45B, GADD45BETA, MYD118 | growth arrest and DNA-damage-inducible, beta |
C00003675
|
| 10912 | GADD45G, CR6, DDIT2, GADD45gamma, GRP17 | growth arrest and DNA-damage-inducible, gamma |
C00003675
|
| 51083 | GAL, GAL-GMAP, GALN, GLNN, GMAP | galanin/GMAP prepropeptide |
C00003675
|
| 374378 | GALNT18, GALNACT18, GALNT15, GALNTL4, GalNAc-T15, GalNAc-T18 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 (EC:2.4.1.41) |
C00003675
|
| 2618 | GART, AIRS, GARS, GARTF, PAIS, PGFT, PRGS | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase (EC:2.1.2.2 6.3.3.1 6.3.4.13) |
C00003675
|
| 283431 | GAS2L3, G2L3 | growth arrest-specific 2 like 3 |
C00003675
|
| 2621 | GAS6, AXLLG, AXSF | growth arrest-specific 6 |
C00003675
|
| 652968 | GATSL3 | GATS protein-like 3 |
C00003675
|
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible |
C00003675
|
| 2634 | GBP2 | guanylate binding protein 2, interferon-inducible |
C00003675
|
| 2635 | GBP3 | guanylate binding protein 3 |
C00003675
|
| 79153 | GDPD3 | glycerophosphodiester phosphodiesterase domain containing 3 |
C00003675
|
| 8487 | GEMIN2, SIP1, SIP1-delta | gem (nuclear organelle) associated protein 2 |
C00003675
|
| 50628 | GEMIN4, HC56, HCAP1, HHRF-1, p97 | gem (nuclear organelle) associated protein 4 |
C00003675
|
| 348654 | GEN1, Gen | GEN1 Holliday junction 5' flap endonuclease |
C00003675
|
| 8836 | GGH, GH | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (EC:3.4.19.9) |
C00003675
|
| 92086 | GGTLC1, GGTL6, GGTLA3, GGTLA4, dJ831C21.1, dJ831C21.2 | gamma-glutamyltransferase light chain 1 (EC:2.3.2.2) |
C00003675
|
| 91227 | GGTLC2, GGTL4 | gamma-glutamyltransferase light chain 2 |
C00003675
|
| 2688 | GH1, GH, GH-N, GHN, IGHD1B, hGH-N | growth hormone 1 |
C00003675
|
| 9837 | GINS1, PSF1 | GINS complex subunit 1 (Psf1 homolog) |
C00003675
|
| 51659 | GINS2, HSPC037, PSF2, Pfs2 | GINS complex subunit 2 (Psf2 homolog) |
C00003675
|
| 64785 | GINS3, PSF3 | GINS complex subunit 3 (Psf3 homolog) |
C00003675
|
| 2706 | GJB2, CX26, DFNA3, DFNA3A, DFNB1, DFNB1A, HID, KID, NSRD1, PPK | gap junction protein, beta 2, 26kDa |
C00003675
|
| 2717 | GLA, GALA | galactosidase, alpha (EC:3.2.1.22) |
C00003675
|
| 152007 | GLIPR2, C9orf19, GAPR-1, GAPR1 | GLI pathogenesis-related 2 |
C00003675
|
| 2746 | GLUD1, GDH, GDH1, GLUD | glutamate dehydrogenase 1 (EC:1.4.1.3) |
C00003675
|
| 2747 | GLUD2, GDH2, GLUDP1 | glutamate dehydrogenase 2 (EC:1.4.1.3) |
C00003675
|
| 51053 | GMNN, Gem | geminin, DNA replication inhibitor |
C00003675
|
| 27232 | GNMT | glycine N-methyltransferase (EC:2.1.1.20) |
C00003675
|
| 10082 | GPC6, OMIMD1 | glypican 6 |
C00003675
|
| 2822 | GPLD1, GPIPLD, GPIPLDM, PIGPLD, PIGPLD1 | glycosylphosphatidylinositol specific phospholipase D1 (EC:3.1.4.50) |
C00003675
|
| 7107 | GPR137B, TM7SF1 | G protein-coupled receptor 137B |
C00003675
|
| 4935 | GPR143, NYS6, OA1 | G protein-coupled receptor 143 |
C00003675
|
| 387509 | GPR153, PGR1 | G protein-coupled receptor 153 |
C00003675
|
| 2842 | GPR19 | G protein-coupled receptor 19 |
C00003675
|
| 9283 | GPR37L1, ET(B)R-LP-2, ETBR-LP-2 | G protein-coupled receptor 37 like 1 |
C00003675
|
| 29899 | GPSM2, CMCS, DFNB82, LGN, PINS | G-protein signaling modulator 2 |
C00003675
|
| 2877 | GPX2, GI-GPx, GPRP, GPRP-2, GPx-2, GPx-GI, GSHPX-GI, GSHPx-2 | glutathione peroxidase 2 (gastrointestinal) (EC:1.11.1.9) |
C00003675
|
| 9687 | GREB1 | growth regulation by estrogen in breast cancer 1 |
C00003675
|
| 2869 | GRK5, GPRK5 | G protein-coupled receptor kinase 5 (EC:2.7.11.16) |
C00003675
|
| 83903 | GSG2, HASPIN | germ cell associated 2 (haspin) (EC:2.7.11.1) |
C00003675
|
| 2932 | GSK3B | glycogen synthase kinase 3 beta (EC:2.7.11.1 2.7.11.26) |
C00003675
|
| 2934 | GSN, ADF, AGEL | gelsolin |
C00003675
|
| 2936 | GSR | glutathione reductase (EC:1.8.1.7) |
C00003675
|
| 2944 | GSTM1, GST1, GSTM1-1, GSTM1a-1a, GSTM1b-1b, GTH4, GTM1, H-B, MU, MU-1 | glutathione S-transferase mu 1 (EC:2.5.1.18) |
C00003675
|
| 2946 | GSTM2, GST4, GSTM, GSTM2-2, GTHMUS | glutathione S-transferase mu 2 (muscle) (EC:2.5.1.18) |
C00003675
|
| 51512 | GTSE1, B99 | G-2 and S-phase expressed 1 |
C00003675
|
| 2997 | GYS1, GSY, GYS | glycogen synthase 1 (muscle) (EC:2.4.1.11) |
C00003675
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00003675
|
| 9563 | H6PD, CORTRD1, G6PDH, GDH | hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (EC:1.1.1.47 3.1.1.31) |
C00003675
|
| 54930 | HAUS4, C14orf94 | HAUS augmin-like complex, subunit 4 |
C00003675
|
| 23354 | HAUS5, KIAA0841, dgt5 | HAUS augmin-like complex, subunit 5 |
C00003675
|
| 55559 | HAUS7, UCHL5IP, UIP1 | HAUS augmin-like complex, subunit 7 |
C00003675
|
| 93323 | HAUS8, DGT4, HICE1, NY-SAR-48 | HAUS augmin-like complex, subunit 8 |
C00003675
|
| 1839 | HBEGF, DTR, DTS, DTSF, HEGFL | heparin-binding EGF-like growth factor |
C00003675
|
| 26959 | HBP1 | HMG-box transcription factor 1 |
C00003675
|
| 79885 | HDAC11, HD11 | histone deacetylase 11 (EC:3.5.1.98) |
C00003675
|
| 51020 | HDDC2, C6orf74, NS5ATP2, dJ167O5.2 | HD domain containing 2 |
C00003675
|
| 57520 | HECW2, NEDL2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
C00003675
|
| 3070 | HELLS, LSH, PASG, SMARCA6 | helicase, lymphoid-specific |
C00003675
|
| 220296 | HEPACAM, GlialCAM, MLC2A, MLC2B | hepatic and glial cell adhesion molecule |
C00003675
|
| 641654 | HEPN1 | hepatocellular carcinoma, down-regulated 1 |
C00003675
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00003675
|
| 54626 | HES2, bHLHb40 | hes family bHLH transcription factor 2 |
C00003675
|
| 79802 | HHIPL2, KIAA1822L | HHIP-like 2 |
C00003675
|
| 26275 | HIBCH, HIBYLCOAH | 3-hydroxyisobutyryl-CoA hydrolase (EC:3.1.2.4) |
C00003675
|
| 28996 | HIPK2, PRO0593 | homeodomain interacting protein kinase 2 (EC:2.7.11.1) |
C00003675
|
| 8479 | HIRIP3 | HIRA interacting protein 3 |
C00003675
|
| 3009 | HIST1H1B, H1, H1.5, H1B, H1F5, H1s-3 | histone cluster 1, H1b |
C00003675
|
| 8332 | HIST1H2AL, H2A.i, H2A/i, H2AFI, dJ193B12.9 | histone cluster 1, H2al |
C00003675
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00003675
|
| 8348 | HIST1H2BO, H2B.2, H2B/n, H2BFN, dJ193B12.2 | histone cluster 1, H2bo |
C00003675
|
| 8350 | HIST1H3A, H3/A, H3FA | histone cluster 1, H3a |
C00003675
|
| 8351 | HIST1H3D, H3/b, H3FB | histone cluster 1, H3d |
C00003675
|
| 8364 | HIST1H4C, H4/g, H4FG, dJ221C16.1 | histone cluster 1, H4c |
C00003675
|
| 8365 | HIST1H4H, H4/h, H4FH | histone cluster 1, H4h |
C00003675
|
| 55355 | HJURP, FAKTS, URLC9, hFLEG1 | Holliday junction recognition protein |
C00003675
|
| 3133 | HLA-E, EA1.2, EA2.1, HLA-6.2, MHC, QA1 | major histocompatibility complex, class I, E |
C00003675
|
| 3131 | HLF | hepatic leukemia factor |
C00003675
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00003675
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00003675
|
| 3157 | HMGCS1, HMGCS | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) (EC:2.3.3.10) |
C00003675
|
| 3161 | HMMR, CD168, IHABP, RHAMM | hyaluronan-mediated motility receptor (RHAMM) |
C00003675
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00003675
|
| 3178 | HNRNPA1, ALS19, ALS20, HNRPA1, HNRPA1L3, IBMPFD3, hnRNP_A1, hnRNP-A1 | heterogeneous nuclear ribonucleoprotein A1 |
C00003675
|
| 3206 | HOXA10, HOX1, HOX1.8, HOX1H, PL | homeobox A10 |
C00003675
|
| 3202 | HOXA5, HOX1, HOX1.3, HOX1C | homeobox A5 |
C00003675
|
| 3248 | HPGD, 15-PGDH, PGDH, PGDH1, PHOAR1, SDR36C1 | hydroxyprostaglandin dehydrogenase 15-(NAD) (EC:1.1.1.141) |
C00003675
|
| 3251 | HPRT1, HGPRT, HPRT | hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8) |
C00003675
|
| 646962 | HRCT1, LGLL338, PRO537, UNQ338 | histidine rich carboxyl terminus 1 |
C00003675
|
| 3290 | HSD11B1, 11-DH, 11-beta-HSD1, CORTRD2, HDL, HSD11, HSD11B, HSD11L, SDR26C1 | hydroxysteroid (11-beta) dehydrogenase 1 (EC:1.1.1.146) |
C00003675
|
| 3291 | HSD11B2, AME, AME1, HSD11K, HSD2, SDR9C3 | hydroxysteroid (11-beta) dehydrogenase 2 (EC:1.1.1.146) |
C00003675
|
| 51170 | HSD17B11, 17BHSD11, DHRS8, PAN1B, RETSDR2, SDR16C2 | hydroxysteroid (17-beta) dehydrogenase 11 (EC:1.1.1.62) |
C00003675
|
| 3294 | HSD17B2, EDH17B2, HSD17, SDR9C2 | hydroxysteroid (17-beta) dehydrogenase 2 (EC:1.1.1.62 1.1.1.239) |
C00003675
|
| 3293 | HSD17B3, EDH17B3, SDR12C2 | hydroxysteroid (17-beta) dehydrogenase 3 (EC:1.1.1.64) |
C00003675
|
| 84941 | HSH2D, ALX, HSH2 | hematopoietic SH2 domain containing |
C00003675
|
| 26353 | HSPB8, CMT2L, DHMN2, E2IG1, H11, HMN2, HMN2A, HSP22 | heat shock 22kDa protein 8 |
C00003675
|
| 3358 | HTR2C, 5-HT2C, 5-HTR2C, HTR1C | 5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
C00003675
|
| 27429 | HTRA2, OMI, PARK13, PRSS25 | HtrA serine peptidase 2 (EC:3.4.21.108) |
C00003675
|
| 3430 | IFI35, IFP35 | interferon-induced protein 35 |
C00003675
|
| 3434 | IFIT1, C56, G10P1, IFI-56, IFI-56K, IFI56, IFIT-1, IFNAI1, ISG56, P56, RNM561 | interferon-induced protein with tetratricopeptide repeats 1 |
C00003675
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00003675
|
| 3481 | IGF2, C11orf43, IGF-II, PP9974 | insulin-like growth factor 2 (somatomedin A) |
C00003675
|
| 3484 | IGFBP1, AFBP, IBP1, IGF-BP25, PP12, hIGFBP-1 | insulin-like growth factor binding protein 1 |
C00003675
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00003675
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00003675
|
| 3489 | IGFBP6, IBP6 | insulin-like growth factor binding protein 6 |
C00003675
|
| 57549 | IGSF9, FP18798, IGSF9A, Nrt1 | immunoglobulin superfamily, member 9 |
C00003675
|
| 9641 | IKBKE, IKK-E, IKK-i, IKKE, IKKI | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon (EC:2.7.11.10) |
C00003675
|
| 100136114 |
C00003675
|
||
| 55540 | IL17RB, CRL4, EVI27, IL17BR, IL17RH1 | interleukin 17 receptor B |
C00003675
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00003675
|
| 3561 | IL2RG, CD132, CIDX, IL-2RG, IMD4, P64, SCIDX, SCIDX1 | interleukin 2 receptor, gamma |
C00003675
|
| 9235 | IL32, IL-32alpha, IL-32beta, IL-32delta, IL-32gamma, NK4, TAIF, TAIFa, TAIFb, TAIFc, TAIFd | interleukin 32 |
C00003675
|
| 3566 | IL4R, CD124, IL-4RA, IL4RA | interleukin 4 receptor |
C00003675
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00003675
|
| 3572 | IL6ST, CD130, CDW130, GP130, IL-6RB | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
C00003675
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00003675
|
| 3619 | INCENP | inner centromere protein antigens 135/155kDa |
C00003675
|
| 3623 | INHA | inhibin, alpha |
C00003675
|
| 3624 | INHBA, EDF, FRP | inhibin, beta A |
C00003675
|
| 3630 | INS, IDDM2, ILPR, IRDN, MODY10 | insulin |
C00003675
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00003675
|
| 51141 | INSIG2 | insulin induced gene 2 |
C00003675
|
| 3640 | INSL3, RLF, RLNL, ley-I-L | insulin-like 3 (Leydig cell) |
C00003675
|
| 3643 | INSR, CD220, HHF5 | insulin receptor (EC:2.7.10.1) |
C00003675
|
| 74918 |
C00003675
|
||
| 128239 | IQGAP3 | IQ motif containing GTPase activating protein 3 |
C00003675
|
| 3665 | IRF7, IRF-7H, IRF7A, IRF7B, IRF7C, IRF7H | interferon regulatory factor 7 |
C00003675
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00003675
|
| 8660 | IRS2, IRS-2 | insulin receptor substrate 2 |
C00003675
|
| 3669 | ISG20, CD25, HEM45 | interferon stimulated exonuclease gene 20kDa (EC:3.1.13.1) |
C00003675
|
| 91464 | ISX, Pix-1, RAXLX | intestine-specific homeobox |
C00003675
|
| 3673 | ITGA2, BR, CD49B, GPIa, HPA-5, VLA-2, VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
C00003675
|
| 3675 | ITGA3, CD49C, GAP-B3, GAPB3, ILNEB, MSK18, VCA-2, VL3A, VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
C00003675
|
| 3694 | ITGB6 | integrin, beta 6 |
C00003675
|
| 80271 | ITPKC, IP3-3KC, IP3KC | inositol-trisphosphate 3-kinase C (EC:2.7.1.127) |
C00003675
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00003675
|
| 389434 | IYD, C6orf71, DEHAL1, TDH4, dJ422F24.1 | iodotyrosine deiodinase (EC:1.22.1.1) |
C00003675
|
| 133746 | JMY, WHDC1L3 | junction mediating and regulatory protein, p53 cofactor |
C00003675
|
| 3751 | KCND2, KV4.2, RK5 | potassium voltage-gated channel, Shal-related subfamily, member 2 |
C00003675
|
| 3752 | KCND3, KCND3L, KCND3S, KSHIVB, KV4.3, SCA19, SCA22 | potassium voltage-gated channel, Shal-related subfamily, member 3 |
C00003675
|
| 3757 | KCNH2, ERG1, HERG, HERG1, Kv11.1, LQT2, SQT1 | potassium voltage-gated channel, subfamily H (eag-related), member 2 |
C00003675
|
| 81033 | KCNH6, ERG-2, ERG2, HERG2, Kv11.2, hERG-2 | potassium voltage-gated channel, subfamily H (eag-related), member 6 |
C00003675
|
| 3758 | KCNJ1, KIR1.1, ROMK, ROMK1 | potassium inwardly-rectifying channel, subfamily J, member 1 |
C00003675
|
| 8645 | KCNK5, K2p5.1, TASK-2, TASK2 | potassium channel, subfamily K, member 5 |
C00003675
|
| 27345 | KCNMB4 | potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
C00003675
|
| 3781 | KCNN2, KCa2.2, SK2, SKCA2, SKCa_2, hSK2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
C00003675
|
| 11015 | KDELR3, ERD2L3 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
C00003675
|
| 84678 | KDM2B, CXXC2, FBXL10, Fbl10, JHDM1B, PCCX2 | lysine (K)-specific demethylase 2B (EC:1.14.11.27) |
C00003675
|
| 9768 | NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | KIAA0101 |
C00003675
|
| 9764 | KIAA0513 |
C00003675
|
|
| 57650 | CIP2A, p90 | KIAA1524 |
C00003675
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00003675
|
| 9928 | KIF14 | kinesin family member 14 |
C00003675
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00003675
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00003675
|
| 146909 | KIF18B | kinesin family member 18B |
C00003675
|
| 23095 | KIF1B, CMT2, CMT2A, CMT2A1, HMSNII, KLP, NBLST1 | kinesin family member 1B |
C00003675
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00003675
|
| 9585 | KIF20B, CT90, KRMP1, MPHOSPH1, MPP-1, MPP1 | kinesin family member 20B |
C00003675
|
| 9493 | KIF23, CHO1, KNSL5, MKLP-1, MKLP1 | kinesin family member 23 |
C00003675
|
| 347240 | KIF24, C9orf48, bA571F15.4 | kinesin family member 24 |
C00003675
|
| 11004 | KIF2C, KNSL6, MCAK | kinesin family member 2C |
C00003675
|
| 24137 | KIF4A, KIF4, KIF4G1 | kinesin family member 4A |
C00003675
|
| 374654 | KIF7, ACLS, HLS2, JBTS12, UNQ340 | kinesin family member 7 |
C00003675
|
| 3833 | KIFC1, HSET, KNSL2 | kinesin family member C1 |
C00003675
|
| 688 | KLF5, BTEB2, CKLF, IKLF | Kruppel-like factor 5 (intestinal) |
C00003675
|
| 3817 | KLK2, KLK2A2, hGK-1, hK2 | kallikrein-related peptidase 2 (EC:3.4.21.35) |
C00003675
|
| 354 | KLK3, APS, KLK2A1, PSA, hK3 | kallikrein-related peptidase 3 (EC:3.4.21.77) |
C00003675
|
| 5653 | KLK6, Bssp, Klk7, PRSS18, PRSS9, SP59, hK6 | kallikrein-related peptidase 6 |
C00003675
|
| 3818 | KLKB1, KLK3, PPK | kallikrein B, plasma (Fletcher factor) 1 (EC:3.4.21.34) |
C00003675
|
| 90417 | KNSTRN, C15orf23, SKAP, TRAF4AF1 | kinetochore-localized astrin/SPAG5 binding protein |
C00003675
|
| 9735 | KNTC1, ROD | kinetochore associated 1 |
C00003675
|
| 779317 |
C00003675
|
||
| 3838 | KPNA2, IPOA1, QIP2, RCH1, SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
C00003675
|
| 83999 | KREMEN1, KREMEN, KRM1 | kringle containing transmembrane protein 1 |
C00003675
|
| 3858 | KRT10, BCIE, BIE, CK10, EHK, K10, KPP | keratin 10 |
C00003675
|
| 3868 | KRT16, CK16, FNEPPK, K16, K1CP, KRT16A, NEPPK | keratin 16 |
C00003675
|
| 3849 | KRT2, CK-2e, K2e, KRT2A, KRT2E, KRTE | keratin 2 |
C00003675
|
| 144501 | KRT80, KB20 | keratin 80 |
C00003675
|
| 3914 | LAMB3, BM600-125KDA, LAM5, LAMNB1 | laminin, beta 3 |
C00003675
|
| 7462 | LAT2, LAB, NTAL, WBSCR15, WBSCR5, WSCR5 | linker for activation of T cells family, member 2 |
C00003675
|
| 3930 | LBR, DHCR14B, LMN2R, PHA, TDRD18 | lamin B receptor |
C00003675
|
| 254251 | LCORL, MLR1 | ligand dependent nuclear receptor corepressor-like |
C00003675
|
| 54741 | LEPROT, LEPR, OB-RGRP, OBRGRP, VPS55 | leptin receptor overlapping transcript |
C00003675
|
| 3955 | LFNG, SCDO3 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (EC:2.4.1.222) |
C00003675
|
| 3965 | LGALS9, HUAT, LGALS9A | lectin, galactoside-binding, soluble, 9 |
C00003675
|
| 654346 | LGALS9C | lectin, galactoside-binding, soluble, 9C |
C00003675
|
| 3972 | LHB, CGB4, LSH-B, hLHB | luteinizing hormone beta polypeptide |
C00003675
|
| 3973 | LHCGR, HHG, LCGR, LGR2, LH/CG-R, LH/CGR, LHR, LHRHR, LSH-R, ULG5 | luteinizing hormone/choriogonadotropin receptor |
C00003675
|
| 10184 | LHFPL2 | lipoma HMGIC fusion partner-like 2 |
C00003675
|
| 89884 | LHX4, CPHD4 | LIM homeobox 4 |
C00003675
|
| 3977 | LIFR, CD118, LIF-R, SJS2, STWS, SWS | leukemia inhibitory factor receptor alpha |
C00003675
|
| 3978 | LIG1 | ligase I, DNA, ATP-dependent (EC:6.5.1.1) |
C00003675
|
| 3987 | LIMS1, PINCH, PINCH-1, PINCH1 | LIM and senescent cell antigen-like domains 1 |
C00003675
|
| 8825 | LIN7A, LIN-7A, LIN7, MALS-1, TIP-33, VELI1 | lin-7 homolog A (C. elegans) |
C00003675
|
| 286826 | LIN9, BARA, BARPsv, Lin-9, TGS, TGS1, TGS2 | lin-9 homolog (C. elegans) |
C00003675
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00003675
|
| 4005 | LMO2, RBTN2, RBTNL1, RHOM2, TTG2 | LIM domain only 2 (rhombotin-like 1) |
C00003675
|
| 80350 | LPAL2, APOA2, APOAL, APOARGC, RP11-72O9.2 | lipoprotein, Lp(a)-like 2, pseudogene |
C00003675
|
| 57913 |
C00003675
|
||
| 57497 | LRFN2, FIGLER2, KIAA1246, RP11-535K1.2, SALM1 | leucine rich repeat and fibronectin type III domain containing 2 |
C00003675
|
| 26018 | LRIG1, LIG-1, LIG1 | leucine-rich repeats and immunoglobulin-like domains 1 |
C00003675
|
| 26020 | LRP10, LRP9, MST087 | low density lipoprotein receptor-related protein 10 |
C00003675
|
| 122769 | LRR1, 4-1BBLRR, LRR-1, PPIL5 | leucine rich repeat protein 1 |
C00003675
|
| 55604 | LRRC16A, CARMIL, CARMIL1, CARMIL1a, LRRC16, dJ501N12.1, dJ501N12.5 | leucine rich repeat containing 16A |
C00003675
|
| 26231 | LRRC29, FBL9, FBXL9 | leucine rich repeat containing 29 |
C00003675
|
| 201255 | LRRC45 | leucine rich repeat containing 45 |
C00003675
|
| 56262 | LRRC8A, AGM5, LRRC8 | leucine rich repeat containing 8 family, member A |
C00003675
|
| 79705 | LRRK1, RIPK6, Roco1 | leucine-rich repeat kinase 1 (EC:2.7.11.1) |
C00003675
|
| 57633 | LRRN1, FIGLER3, NLRR-1 | leucine rich repeat neuronal 1 |
C00003675
|
| 4047 | LSS, OSC | lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (EC:5.4.99.7) |
C00003675
|
| 338645 | LUZP2, KFSP2566, PRO6246 | leucine zipper protein 2 |
C00003675
|
| 56925 | LXN, ECI, TCI | latexin |
C00003675
|
| 23643 | LY96, ESOP-1, MD-2, MD2, ly-96 | lymphocyte antigen 96 |
C00003675
|
| 55646 | LYAR, ZC2HC2, ZLYAR | Ly1 antibody reactive |
C00003675
|
| 4085 | MAD2L1, HSMAD2, MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) |
C00003675
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00003675
|
| 10892 | MALT1, IMD12, MLT, MLT1 | mucosa associated lymphoid tissue lymphoma translocation gene 1 (EC:3.4.22.-) |
C00003675
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00003675
|
| 4294 | MAP3K10, MEKK10, MLK2, MST | mitogen-activated protein kinase kinase kinase 10 (EC:2.7.11.25) |
C00003675
|
| 9020 | MAP3K14, FTDCR1B, HS, HSNIK, NIK | mitogen-activated protein kinase kinase kinase 14 (EC:2.7.11.25) |
C00003675
|
| 256714 | MAP7D2 | MAP7 domain containing 2 |
C00003675
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00003675
|
| 5600 | MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2, p38Beta | mitogen-activated protein kinase 11 (EC:2.7.11.24) |
C00003675
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00003675
|
| 441061 | MARCH11, MARCH-XI | membrane-associated ring finger (C3HC4) 11 (EC:6.3.2.-) |
C00003675
|
| 375449 | MAST4 | microtubule associated serine/threonine kinase family member 4 (EC:2.7.11.1) |
C00003675
|
| 4148 | MATN3, DIPOA, EDM5, HOA, OADIP, OS2 | matrilin 3 |
C00003675
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00003675
|
| 4171 | MCM2, BM28, CCNL1, CDCL1, D3S3194, MITOTIN, cdc19 | minichromosome maintenance complex component 2 (EC:3.6.4.12) |
C00003675
|
| 4172 | MCM3, HCC5, P1-MCM3, P1.h, RLFB | minichromosome maintenance complex component 3 (EC:3.6.4.12) |
C00003675
|
| 4173 | MCM4, CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 | minichromosome maintenance complex component 4 (EC:3.6.4.12) |
C00003675
|
| 4174 | MCM5, CDC46, P1-CDC46 | minichromosome maintenance complex component 5 (EC:3.6.4.12) |
C00003675
|
| 4175 | MCM6, MCG40308, Mis5, P105MCM | minichromosome maintenance complex component 6 (EC:3.6.4.12) |
C00003675
|
| 4176 | MCM7, CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146 | minichromosome maintenance complex component 7 (EC:3.6.4.12) |
C00003675
|
| 84515 | MCM8, C20orf154, dJ967N21.5 | minichromosome maintenance complex component 8 (EC:3.6.4.12) |
C00003675
|
| 79892 | MCMBP, C10orf119, MCM-BP | minichromosome maintenance complex binding protein |
C00003675
|
| 9656 | MDC1, NFBD1 | mediator of DNA-damage checkpoint 1 |
C00003675
|
| 1953 | MEGF6, EGFL3 | multiple EGF-like-domains 6 |
C00003675
|
| 9833 | MELK, HPK38 | maternal embryonic leucine zipper kinase (EC:2.7.11.1 2.7.10.2) |
C00003675
|
| 10461 | MERTK, MER, RP38, c-mer | c-mer proto-oncogene tyrosine kinase (EC:2.7.10.1) |
C00003675
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00003675
|
| 79828 | METTL8, TIP | methyltransferase like 8 |
C00003675
|
| 92312 | MEX3A, MEX-3A, RKHD4 | mex-3 RNA binding family member A |
C00003675
|
| 54842 | MFSD6, MMR2, hMMR2 | major facilitator superfamily domain containing 6 |
C00003675
|
| 11343 | MGLL, HU-K5, HUK5, MAGL, MGL | monoglyceride lipase (EC:3.1.1.23) |
C00003675
|
| 92667 | MGME1, C20orf72, DDK1, MTDPS11, bA504H3.4 | mitochondrial genome maintenance exonuclease 1 |
C00003675
|
| 64780 | MICAL1, MICAL, MICAL-1, NICAL | microtubule associated monooxygenase, calponin and LIM domain containing 1 |
C00003675
|
| 440574 | MINOS1, C1orf151, MIO10 | mitochondrial inner membrane organizing system 1 |
C00003675
|
| 9562 | MINPP1, HIPER1, MINPP2, MIPP | multiple inositol-polyphosphate phosphatase 1 (EC:3.1.3.62 3.1.3.80) |
C00003675
|
| 100302213 | MIR1181, MIRN1181, hsa-mir-1181 | microRNA 1181 |
C00003675
|
| 406924 | MIR134, MIRN134 | microRNA 134 |
C00003675
|
| 100314280 |
C00003675
|
||
| 406952 | MIR17, MIR17-5p, MIR91, MIRN17, MIRN91, hsa-mir-17, miR-17, miR17-3p, miRNA17, miRNA91 | microRNA 17 |
C00003675
|
| 100302129 | MIR1915, MIRN1915, hsa-mir-1915 | microRNA 1915 |
C00003675
|
| 406982 | MIR20A, MIR20, MIRN20, MIRN20A, hsa-mir-20, hsa-mir-20a, miR-20, miRNA20A | microRNA 20a |
C00003675
|
| 574032 | MIR20B, MIRN20B, hsa-mir-20b | microRNA 20b |
C00003675
|
| 407004 | MIR22, MIRN22, hsa-mir-22, miR-22 | microRNA 22 |
C00003675
|
| 84981 | MIR22HG, C17orf91 | MIR22 host gene (non-protein coding) |
C00003675
|
| 407021 | MIR29A, MIRN29, MIRN29A, hsa-mir-29, hsa-mir-29a, miRNA29A | microRNA 29a |
C00003675
|
| 100526320 |
C00003675
|
||
| 81571 | MIR600HG, C9orf45, NCRNA00287 | MIR600 host gene (non-protein coding) |
C00003675
|
| 693214 | MIR629, MIRN629, hsa-mir-629 | microRNA 629 |
C00003675
|
| 54069 | MIS18A, B28, C21orf45, C21orf46, FASP1, MIS18alpha, hMis18alpha | MIS18 kinetochore protein A |
C00003675
|
| 55320 | MIS18BP1, C14orf106, HSA242977, KNL2, M18BP1 | MIS18 binding protein 1 |
C00003675
|
| 4288 | MKI67, KIA, MIB-1 | marker of proliferation Ki-67 |
C00003675
|
| 4289 | MKLN1, TWA2 | muskelin 1, intracellular mediator containing kelch motifs |
C00003675
|
| 54903 | MKS1, BBS13, MES, MKS, POC12 | Meckel syndrome, type 1 |
C00003675
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00003675
|
| 4321 | MMP12, HME, ME, MME, MMP-12 | matrix metallopeptidase 12 (macrophage elastase) (EC:3.4.24.65) |
C00003675
|
| 4325 | MMP16, C8orf57, MMP-X2, MT-MMP2, MT-MMP3, MT3-MMP | matrix metallopeptidase 16 (membrane-inserted) (EC:3.4.24.-) |
C00003675
|
| 253714 | MMS22L, C6orf167, dJ39B17.2 | MMS22-like, DNA repair protein |
C00003675
|
| 84057 | MND1, GAJ | meiotic nuclear divisions 1 homolog (S. cerevisiae) |
C00003675
|
| 55329 | MNS1, SPATA40 | meiosis-specific nuclear structural 1 |
C00003675
|
| 80168 | MOGAT2, DGAT2L5, MGAT2 | monoacylglycerol O-acyltransferase 2 (EC:2.3.1.22) |
C00003675
|
| 23164 | MPRIP, M-RIP, MRIP, RHOIP3, RIP3, p116Rip | myosin phosphatase Rho interacting protein |
C00003675
|
| 4439 | MSH5, G7, MUTSH5, NG23 | mutS homolog 5 |
C00003675
|
| 253827 | MSRB3, DFNB74 | methionine sulfoxide reductase B3 (EC:1.8.4.-) |
C00003675
|
| 4501 | MT1X, MT-1l, MT1 | metallothionein 1X |
C00003675
|
| 27085 | MTBP, MDM2BP | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
C00003675
|
| 113115 | MTFR2, DUFD1, FAM54A | mitochondrial fission regulator 2 |
C00003675
|
| 4524 | MTHFR | methylenetetrahydrofolate reductase (NAD(P)H) (EC:1.5.1.20) |
C00003675
|
| 4544 | MTNR1B, FGQTL2, MEL-1B-R, MT2 | melatonin receptor 1B |
C00003675
|
| 222166 | MTURN, C7orf41, Ells1 | maturin, neural progenitor differentiation regulator homolog (Xenopus) |
C00003675
|
| 9961 | MVP, LRP, VAULT1 | major vault protein |
C00003675
|
| 83463 | MXD3, BHLHC13, MAD3, MYX | MAX dimerization protein 3 |
C00003675
|
| 4602 | MYB, Cmyb, c-myb, c-myb_CDS, efg | v-myb avian myeloblastosis viral oncogene homolog |
C00003675
|
| 4605 | MYBL2, B-MYB, BMYB | v-myb avian myeloblastosis viral oncogene homolog-like 2 |
C00003675
|
| 4604 | MYBPC1, LCCS4, MYBPCC, MYBPCS | myosin binding protein C, slow type |
C00003675
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00003675
|
| 4610 | MYCL, LMYC, MYCL1, bHLHe38 | v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
C00003675
|
| 58498 | MYL7, MYL2A, MYLC2A | myosin, light chain 7, regulatory |
C00003675
|
| 10398 | MYL9, LC20, MLC-2C, MLC2, MRLC1, MYRL2 | myosin, light chain 9, regulatory |
C00003675
|
| 4638 | MYLK, AAT7, KRP, MLCK, MLCK1, MLCK108, MLCK210, MSTP083, MYLK1, smMLCK | myosin light chain kinase (EC:2.7.11.18) |
C00003675
|
| 55892 | MYNN, OSZF, ZBTB31, ZNF902 | myoneurin |
C00003675
|
| 80179 | MYO19, MYOHD1 | myosin XIX |
C00003675
|
| 4642 | MYO1D, myr4 | myosin ID |
C00003675
|
| 4645 | MYO5B | myosin VB |
C00003675
|
| 440145 | MZT1, C13orf37, MOZART1 | mitotic spindle organizing protein 1 |
C00003675
|
| 10135 | NAMPT, 1110035O14Rik, PBEF, PBEF1, VF, VISFATIN | nicotinamide phosphoribosyltransferase (EC:2.4.2.12) |
C00003675
|
| 266812 | NAP1L5, DRLM | nucleosome assembly protein 1-like 5 |
C00003675
|
| 4678 | NASP, FLB7527, PRO1999 | nuclear autoantigenic sperm protein (histone-binding) |
C00003675
|
| 9 | NAT1, AAC1, MNAT, NAT-1, NATI | N-acetyltransferase 1 (arylamine N-acetyltransferase) (EC:2.3.1.5) |
C00003675
|
| 10 | NAT2, AAC2, NAT-2, PNAT | N-acetyltransferase 2 (arylamine N-acetyltransferase) (EC:2.3.1.5) |
C00003675
|
| 89795 | NAV3, POMFIL1, STEERIN3, unc53H3 | neuron navigator 3 |
C00003675
|
| 4681 | NBL1, D1S1733E, DAN, DAND1, NB, NO3 | neuroblastoma 1, DAN family BMP antagonist |
C00003675
|
| 9918 | NCAPD2, CAP-D2, CNAP1, hCAP-D2 | non-SMC condensin I complex, subunit D2 |
C00003675
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00003675
|
| 54892 | NCAPG2, CAP-G2, CAPG2, LUZP5, MTB, hCAP-G2 | non-SMC condensin II complex, subunit G2 |
C00003675
|
| 23397 | NCAPH, BRRN1, CAP-H | non-SMC condensin I complex, subunit H |
C00003675
|
| 54820 | NDE1, HOM-TES-87, LIS4, NDE, NUDE, NUDE1 | nudE neurodevelopment protein 1 |
C00003675
|
| 55247 | NEIL3, FGP2, FPG2, NEI3, hFPG2, hNEI3 | nei endonuclease VIII-like 3 (E. coli) (EC:4.2.99.18) |
C00003675
|
| 4751 | NEK2, HsPK21, NEK2A, NLK1 | NIMA-related kinase 2 (EC:2.7.11.1) |
C00003675
|
| 81832 | NETO1, BCTL1, BTCL1 | neuropilin (NRP) and tolloid (TLL)-like 1 |
C00003675
|
| 60491 | NIF3L1, ALS2CR1, CALS-7 | NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
C00003675
|
| 4815 | NINJ2 | ninjurin 2 |
C00003675
|
| 84166 | NLRC5, CLR16.1, NOD27, NOD4 | NLR family, CARD domain containing 5 |
C00003675
|
| 126206 | NLRP5, CLR19.8, MATER, NALP5, PAN11, PYPAF8 | NLR family, pyrin domain containing 5 |
C00003675
|
| 23057 | NMNAT2, C1orf15, PNAT2 | nicotinamide nucleotide adenylyltransferase 2 (EC:2.7.7.1 2.7.7.18) |
C00003675
|
| 10874 | NMU | neuromedin U |
C00003675
|
| 51602 | NOP58, NOP5, NOP5/NOP58 | NOP58 ribonucleoprotein |
C00003675
|
| 4842 | NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS | nitric oxide synthase 1 (neuronal) (EC:1.14.13.39) |
C00003675
|
| 4843 | NOS2, HEP-NOS, INOS, NOS, NOS2A | nitric oxide synthase 2, inducible (EC:1.14.13.39) |
C00003675
|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00003675
|
| 27035 | NOX1, GP91-2, MOX1, NOH-1, NOH1 | NADPH oxidase 1 |
C00003675
|
| 4869 | NPM1, B23, NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
C00003675
|
| 4880 | NPPC, CNP, CNP2 | natriuretic peptide C |
C00003675
|
| 126382 | NR2C2AP, TRA16 | nuclear receptor 2C2-associated protein |
C00003675
|
| 4306 | NR3C2, MCR, MLR, MR, NR3C2VIT | nuclear receptor subfamily 3, group C, member 2 |
C00003675
|
| 3164 | NR4A1, GFRP1, HMR, N10, NAK-1, NGFIB, NP10, NUR77, TR3 | nuclear receptor subfamily 4, group A, member 1 |
C00003675
|
| 11270 | NRM, NRM29 | nurim (nuclear envelope membrane protein) |
C00003675
|
| 84628 | NTNG2, LHLL9381, Lmnt2, NTNG1, bA479K20.1 | netrin G2 |
C00003675
|
| 4925 | NUCB2, NEFA | nucleobindin 2 |
C00003675
|
| 4521 | NUDT1, MTH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 (EC:3.6.1.55 3.6.1.56) |
C00003675
|
| 79873 | NUDT18, MTH3 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 (EC:3.6.1.58) |
C00003675
|
| 83540 | NUF2, CDCA1, CT106, NUF2R | NUF2, NDC80 kinetochore complex component |
C00003675
|
| 57122 | NUP107, NUP84 | nucleoporin 107kDa |
C00003675
|
| 9631 | NUP155, N155 | nucleoporin 155kDa |
C00003675
|
| 129401 | NUP35, MP44, NP44, NUP53 | nucleoporin 35kDa |
C00003675
|
| 79023 | NUP37, p37 | nucleoporin 37kDa |
C00003675
|
| 53371 | NUP54 | nucleoporin 54kDa |
C00003675
|
| 54830 | NUP62CL | nucleoporin 62kDa C-terminal like |
C00003675
|
| 51203 | NUSAP1, ANKT, BM037, LNP, NUSAP, PRO0310p1, Q0310, SAPL | nucleolar and spindle associated protein 1 |
C00003675
|
| 55998 | NXF5 | nuclear RNA export factor 5 |
C00003675
|
| 4938 | OAS1, IFI-4, OIAS, OIASI | 2'-5'-oligoadenylate synthetase 1, 40/46kDa (EC:2.7.7.84) |
C00003675
|
| 4940 | OAS3, p100 | 2'-5'-oligoadenylate synthetase 3, 100kDa (EC:2.7.7.84) |
C00003675
|
| 79629 | OCEL1, FWP009, S863-9 | occludin/ELL domain containing 1 |
C00003675
|
| 54959 | ODAM, APIN | odontogenic, ameloblast asssociated |
C00003675
|
| 11339 | OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta, hMIS18beta | Opa interacting protein 5 |
C00003675
|
| 55301 | OLAH, AURA1, SAST, THEDC1 | oleoyl-ACP hydrolase (EC:3.1.2.14) |
C00003675
|
| 4983 | OPHN1, ARHGAP41, MRX60, OPN1 | oligophrenin 1 |
C00003675
|
| 10133 | OPTN, ALS12, FIP2, GLC1E, HIP7, HYPL, NRP, TFIIIA-INTP | optineurin |
C00003675
|
| 4998 | ORC1, HSORC1, ORC1L, PARC1 | origin recognition complex, subunit 1 |
C00003675
|
| 716175 |
C00003675
|
||
| 5004 | ORM1, AGP-A, AGP1, ORM | orosomucoid 1 |
C00003675
|
| 5005 | ORM2, AGP-B, AGP-B', AGP2 | orosomucoid 2 |
C00003675
|
| 114876 | OSBPL1A, ORP-1, ORP1, OSBPL1B | oxysterol binding protein-like 1A |
C00003675
|
| 114881 | OSBPL7, ORP7 | oxysterol binding protein-like 7 |
C00003675
|
| 29948 | OSGIN1, BDGI, OKL38 | oxidative stress induced growth inhibitor 1 |
C00003675
|
| 116039 | OSR2 | odd-skipped related transciption factor 2 |
C00003675
|
| 56957 | OTUD7B, CEZANNE, ZA20D1 | OTU domain containing 7B (EC:3.4.19.12) |
C00003675
|
| 55690 | PACS1, MRD17 | phosphofurin acidic cluster sorting protein 1 |
C00003675
|
| 29993 | PACSIN1, SDPI | protein kinase C and casein kinase substrate in neurons 1 |
C00003675
|
| 5047 | PAEP, GD, GdA, GdF, GdS, PAEG, PEP, PP14 | progestagen-associated endometrial protein |
C00003675
|
| 55003 | PAK1IP1, MAK11, PIP1, RP11-421M1.5, WDR84, bA421M1.5, hPIP1 | PAK1 interacting protein 1 |
C00003675
|
| 10038 | PARP2, ADPRT2, ADPRTL2, ADPRTL3, ARTD2, PARP-2, pADPRT-2 | poly (ADP-ribose) polymerase 2 (EC:2.4.2.30) |
C00003675
|
| 55010 | PARPBP, AROM, C12orf48, PARI | PARP1 binding protein |
C00003675
|
| 5075 | PAX1, HUP48 | paired box 1 |
C00003675
|
| 55872 | PBK, CT84, Nori-3, SPK, TOPK | PDZ binding kinase (EC:2.7.12.2) |
C00003675
|
| 5097 | PCDH1, PC42, PCDH42 | protocadherin 1 |
C00003675
|
| 56110 | PCDHGA5, CDH-GAMMA-A5, ME3, PCDH-GAMMA-A5 | protocadherin gamma subfamily A, 5 |
C00003675
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00003675
|
| 5046 | PCSK6, PACE4, SPC4 | proprotein convertase subtilisin/kexin type 6 |
C00003675
|
| 58488 | PCTP, PC-TP, STARD2 | phosphatidylcholine transfer protein |
C00003675
|
| 5139 | PDE3A, CGI-PDE, CGI-PDE_A, CGI-PDE-A | phosphodiesterase 3A, cGMP-inhibited (EC:3.1.4.17) |
C00003675
|
| 5142 | PDE4B, DPDE4, PDE4B5, PDEIVB | phosphodiesterase 4B, cAMP-specific (EC:3.1.4.17) |
C00003675
|
| 5165 | PDK3, CMTX6 | pyruvate dehydrogenase kinase, isozyme 3 (EC:2.7.11.2) |
C00003675
|
| 64236 | PDLIM2, MYSTIQUE, SLIM | PDZ and LIM domain 2 (mystique) |
C00003675
|
| 23590 | PDSS1, COQ1, COQ10D2, DPS, RP13-16H11.3, SPS, TPRT, TPT, TPT_1, hDPS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 (EC:2.5.1.91) |
C00003675
|
| 10158 | PDZK1IP1, DD96, MAP17, RP1-18D14.5, SPAP | PDZK1 interacting protein 1 |
C00003675
|
| 157310 | PEBP4, CORK-1, CORK1, GWTM1933, PEBP-4, PRO4408, hPEBP4 | phosphatidylethanolamine-binding protein 4 |
C00003675
|
| 5194 | PEX13, NALD, PBD11A, PBD11B, ZWS | peroxisomal biogenesis factor 13 |
C00003675
|
| 5198 | PFAS, FGAMS, FGARAT, PURL | phosphoribosylformylglycinamidine synthase (EC:6.3.5.3) |
C00003675
|
| 5208 | PFKFB2, PFK-2/FBPase-2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (EC:2.7.1.105 3.1.3.46) |
C00003675
|
| 5225 | PGC, PEPC, PGII | progastricsin (pepsinogen C) (EC:3.4.23.3) |
C00003675
|
| 5238 | PGM3, AGM1, PAGM, PGM_3 | phosphoglucomutase 3 (EC:5.4.2.3) |
C00003675
|
| 54858 | PGPEP1, PGP, PGP-I, PGPI, Pcp | pyroglutamyl-peptidase I (EC:3.4.19.3) |
C00003675
|
| 5252 | PHF1, MTF2L2, PCL1, PHF2, TDRD19C, hPHF1 | PHD finger protein 1 |
C00003675
|
| 26147 | PHF19, MTF2L1, PCL3, TDRD19B | PHD finger protein 19 |
C00003675
|
| 644844 | PHGR1 | proline/histidine/glycine-rich 1 |
C00003675
|
| 57157 | PHTF2 | putative homeodomain transcription factor 2 |
C00003675
|
| 85007 | PHYKPL, AGXT2L2, PHLU | 5-phosphohydroxy-L-lysine phospho-lyase (EC:4.2.3.134) |
C00003675
|
| 8554 | PIAS1, DDXBP1, GBP, GU/RH-II, ZMIZ3 | protein inhibitor of activated STAT, 1 |
C00003675
|
| 80119 | PIF1, C15orf20, PIF | PIF1 5'-to-3' DNA helicase (EC:3.6.4.12) |
C00003675
|
| 284098 | PIGW, Gwt1 | phosphatidylinositol glycan anchor biosynthesis, class W |
C00003675
|
| 113791 | PIK3IP1, HGFL, hHGFL(S) | phosphoinositide-3-kinase interacting protein 1 |
C00003675
|
| 9088 | PKMYT1, MYT1 | protein kinase, membrane associated tyrosine/threonine 1 (EC:2.7.11.1) |
C00003675
|
| 63876 | PKNOX2, PREP2 | PBX/knotted 1 homeobox 2 |
C00003675
|
| 5318 | PKP2, ARVD9 | plakophilin 2 |
C00003675
|
| 8502 | PKP4, p0071 | plakophilin 4 |
C00003675
|
| 10761 | PLAC1, CT92, OOSP2L | placenta-specific 1 |
C00003675
|
| 5329 | PLAUR, CD87, U-PAR, UPAR, URKR | plasminogen activator, urokinase receptor |
C00003675
|
| 113026 | PLCD3 | phospholipase C, delta 3 (EC:3.1.4.11) |
C00003675
|
| 23007 | PLCH1, PLCL3 | phospholipase C, eta 1 (EC:3.1.4.11) |
C00003675
|
| 5338 | PLD2 | phospholipase D2 (EC:3.1.4.4) |
C00003675
|
| 22874 | PLEKHA6, PEPP-3, PEPP3 | pleckstrin homology domain containing, family A member 6 |
C00003675
|
| 55041 | PLEKHB2, EVT2 | pleckstrin homology domain containing, family B (evectins) member 2 |
C00003675
|
| 55200 | PLEKHG6, MyoGEF | pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
C00003675
|
| 5347 | PLK1, PLK, STPK13 | polo-like kinase 1 (EC:2.7.11.21) |
C00003675
|
| 10733 | PLK4, SAK, STK18 | polo-like kinase 4 (EC:2.7.11.21) |
C00003675
|
| 51090 | PLLP, PMLP, TM4SF11 | plasmolipin |
C00003675
|
| 55558 | PLXNA3, 6.3, HSSEXGENE, PLXN3, PLXN4, XAP-6 | plexin A3 |
C00003675
|
| 5367 | PMCH, MCH | pro-melanin-concentrating hormone |
C00003675
|
| 5406 | PNLIP, PL, PNLIPD, PTL | pancreatic lipase (EC:3.1.1.3) |
C00003675
|
| 25886 | POC1A, PIX2, SOFT, WDR51A | POC1 centriolar protein A |
C00003675
|
| 5420 | PODXL, Gp200, PC, PCLP, PCLP-1 | podocalyxin-like |
C00003675
|
| 23649 | POLA2 | polymerase (DNA directed), alpha 2, accessory subunit |
C00003675
|
| 5424 | POLD1, CDC2, CRCS10, MDPL, POLD | polymerase (DNA directed), delta 1, catalytic subunit (EC:2.7.7.7) |
C00003675
|
| 5427 | POLE2, DPE2 | polymerase (DNA directed), epsilon 2, accessory subunit (EC:2.7.7.7) |
C00003675
|
| 10721 | POLQ, POLH, PRO0327 | polymerase (DNA directed), theta (EC:2.7.7.7) |
C00003675
|
| 10622 | POLR3G, RPC32, RPC7 | polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
C00003675
|
| 721804 |
C00003675
|
||
| 100996331 | POTEB, A26B1, CT104.5, POTE-15, POTE15 | POTE ankyrin domain family, member B |
C00003675
|
| 388468 | POTEC, A26B2, CT104.6, POTE-18, POTE18 | POTE ankyrin domain family, member C |
C00003675
|
| 445582 | POTEE, A26C1, A26C1A, CT104.2, POTE-2, POTE2, POTE2gamma | POTE ankyrin domain family, member E |
C00003675
|
| 23784 | POTEH, A26C3, ACTBL1, CT104.7, POTE22 | POTE ankyrin domain family, member H |
C00003675
|
| 641455 | POTEM, ACT, P704P, POTE14beta | POTE ankyrin domain family, member M |
C00003675
|
| 5465 | PPARA, NR1C1, PPAR, PPARalpha, hPPAR | peroxisome proliferator-activated receptor alpha |
C00003675
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00003675
|
| 8495 | PPFIBP2, Cclp1 | PTPRF interacting protein, binding protein 2 (liprin beta 2) |
C00003675
|
| 5480 | PPIC, CYPC | peptidylprolyl isomerase C (cyclophilin C) (EC:5.2.1.8) |
C00003675
|
| 10465 | PPIH, CYP-20, CYPH, SnuCyp-20, USA-CYP | peptidylprolyl isomerase H (cyclophilin H) (EC:5.2.1.8) |
C00003675
|
| 5493 | PPL | periplakin |
C00003675
|
| 5500 | PPP1CB, PP-1B, PP1B, PP1beta, PPP1CD | protein phosphatase 1, catalytic subunit, beta isozyme (EC:3.1.3.16 3.1.3.53) |
C00003675
|
| 81706 | PPP1R14C, CPI17-like, KEPI, NY-BR-81 | protein phosphatase 1, regulatory (inhibitor) subunit 14C |
C00003675
|
| 28227 | PPP2R3B, NYREN8, PPP2R3L, PPP2R3LY, PR48 | protein phosphatase 2, regulatory subunit B'', beta |
C00003675
|
| 5526 | PPP2R5B, B56B, PR61B | protein phosphatase 2, regulatory subunit B', beta |
C00003675
|
| 54896 | PQLC2 | PQ loop repeat containing 2 |
C00003675
|
| 9055 | PRC1, ASE1 | protein regulator of cytokinesis 1 |
C00003675
|
| 5557 | PRIM1, p49 | primase, DNA, polypeptide 1 (49kDa) (EC:2.7.7.7) |
C00003675
|
| 145270 | PRIMA1, PRIMA | proline rich membrane anchor 1 |
C00003675
|
| 201973 | PRIMPOL, CCDC111, MYP22 | primase and polymerase (DNA-directed) |
C00003675
|
| 5562 | PRKAA1, AMPK, AMPKa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit (EC:2.7.11.1 2.7.11.26 2.7.11.27 2.7.11.31) |
C00003675
|
| 5587 | PRKD1, PKC-MU, PKCM, PKD, PRKCM | protein kinase D1 (EC:2.7.11.13) |
C00003675
|
| 5617 | PRL | prolactin |
C00003675
|
| 5625 | PRODH, HSPOX2, PIG6, POX, PRODH1, PRODH2, TP53I6 | proline dehydrogenase (oxidase) 1 (EC:1.5.99.8) |
C00003675
|
| 254427 | PROSER2, C10orf47 | proline and serine-rich protein 2 |
C00003675
|
| 84950 | PRPF38A, Prp38, RP5-965L7.1 | pre-mRNA processing factor 38A |
C00003675
|
| 55771 | PRR11 | proline rich 11 |
C00003675
|
| 51334 | PRR16, DSC54 | proline rich 16 |
C00003675
|
| 80742 | PRR3, CAT56 | proline rich 3 |
C00003675
|
| 64063 | PRSS22, BSSP-4, hBSSP-4 | protease, serine, 22 |
C00003675
|
| 11098 | PRSS23, SIG13, SPUVE, ZSIG13 | protease, serine, 23 |
C00003675
|
| 5699 | PSMB10, LMP10, MECL1, beta2i | proteasome (prosome, macropain) subunit, beta type, 10 (EC:3.4.25.1) |
C00003675
|
| 29893 | PSMC3IP, GT198, HOP2, HUMGT198A, ODG3, TBPIP | PSMC3 interacting protein |
C00003675
|
| 5714 | PSMD8, HIP6, HYPF, Nin1p, Rpn12, S14, p31 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
C00003675
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00003675
|
| 5744 | PTHLH, BDE2, HHM, PLP, PTHR, PTHRP | parathyroid hormone-like hormone |
C00003675
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00003675
|
| 11099 | PTPN21, PTPD1, PTPRL10 | protein tyrosine phosphatase, non-receptor type 21 (EC:3.1.3.48) |
C00003675
|
| 5797 | PTPRM, PTPRL1, R-PTP-MU, RPTPM, RPTPU, hR-PTPu | protein tyrosine phosphatase, receptor type, M (EC:3.1.3.48) |
C00003675
|
| 5799 | PTPRN2, IA-2beta, IAR, ICAAR, PTPRP, R-PTP-N2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 (EC:3.1.3.48) |
C00003675
|
| 284119 | PTRF, CAVIN, CAVIN1, CGL4, cavin-1 | polymerase I and transcript release factor |
C00003675
|
| 9232 | PTTG1, EAP1, HPTTG, PTTG, TUTR1 | pituitary tumor-transforming 1 |
C00003675
|
| 10744 | PTTG2 | pituitary tumor-transforming 2 |
C00003675
|
| 5768 | QSOX1, Q6, QSCN6 | quiescin Q6 sulfhydryl oxidase 1 (EC:1.8.3.2) |
C00003675
|
| 80223 | RAB11FIP1, NOEL1A, RCP, rab11-FIP1 | RAB11 family interacting protein 1 (class I) |
C00003675
|
| 51715 | RAB23, HSPC137 | RAB23, member RAS oncogene family |
C00003675
|
| 53916 | RAB4B | RAB4B, member RAS oncogene family |
C00003675
|
| 29127 | RACGAP1, CYK4, HsCYK-4, ID-GAP, MgcRacGAP | Rac GTPase activating protein 1 |
C00003675
|
| 56852 | RAD18, RNF73 | RAD18 homolog (S. cerevisiae) |
C00003675
|
| 5888 | RAD51, BRCC5, HRAD51, HsRad51, HsT16930, MRMV2, RAD51A, RECA | RAD51 recombinase |
C00003675
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00003675
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00003675
|
| 8438 | RAD54L, HR54, RAD54A, hHR54, hRAD54 | RAD54-like (S. cerevisiae) |
C00003675
|
| 5894 | RAF1, CRAF, NS5, Raf-1, c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 (EC:2.7.11.1) |
C00003675
|
| 26064 | RAI14, NORPEG, RAI13 | retinoic acid induced 14 |
C00003675
|
| 10742 | RAI2 | retinoic acid induced 2 |
C00003675
|
| 57186 | RALGAPA2, AS250, C20orf74, bA287B20.1, dJ1049G11, dJ1049G11.4, p220 | Ral GTPase activating protein, alpha subunit 2 (catalytic) |
C00003675
|
| 10267 | RAMP1 | receptor (G protein-coupled) activity modifying protein 1 |
C00003675
|
| 65059 | RAPH1, ALS2CR18, ALS2CR9, LPD, PREL-2, PREL2, RMO1, RalGDS/AF-6 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
C00003675
|
| 5920 | RARRES3, HRASLS4, HRSL4, PLA1/2-3, RIG1, TIG3 | retinoic acid receptor responder (tazarotene induced) 3 |
C00003675
|
| 51655 | RASD1, AGS1, DEXRAS1, MGC:26290 | RAS, dexamethasone-induced 1 |
C00003675
|
| 387496 | RASL11A | RAS-like, family 11, member A |
C00003675
|
| 65997 | RASL11B | RAS-like, family 11, member B |
C00003675
|
| 5932 | RBBP8, COM1, CTIP, JWDS, RIM, SAE2, SCKL2 | retinoblastoma binding protein 8 |
C00003675
|
| 5933 | RBL1, CP107, PRB1, p107 | retinoblastoma-like 1 (p107) |
C00003675
|
| 494115 | RBMXL1, RBM1 | RNA binding motif protein, X-linked-like 1 |
C00003675
|
| 1827 | RCAN1, ADAPT78, CSP1, DSC1, DSCR1, MCIP1, RCN1 | regulator of calcineurin 1 |
C00003675
|
| 91433 | RCCD1 | RCC1 domain containing 1 |
C00003675
|
| 201299 | RDM1, RAD52B | RAD52 motif 1 |
C00003675
|
| 9401 | RECQL4, RECQ4 | RecQ protein-like 4 (EC:3.6.4.12) |
C00003675
|
| 5971 | RELB, I-REL, IREL, REL-B | v-rel avian reticuloendotheliosis viral oncogene homolog B |
C00003675
|
| 5982 | RFC2, RFC40 | replication factor C (activator 1) 2, 40kDa |
C00003675
|
| 5983 | RFC3, RFC38 | replication factor C (activator 1) 3, 38kDa |
C00003675
|
| 5984 | RFC4, A1, RFC37 | replication factor C (activator 1) 4, 37kDa |
C00003675
|
| 5988 | RFPL1, RNF78 | ret finger protein-like 1 |
C00003675
|
| 55159 | RFWD3, RNF201 | ring finger and WD repeat domain 3 |
C00003675
|
| 285704 | RGMB, DRAGON | repulsive guidance molecule family member b |
C00003675
|
| 8786 | RGS11, RS11 | regulator of G-protein signaling 11 |
C00003675
|
| 26575 | RGS17, RGS-17, RGSZ2, hRGS17 | regulator of G-protein signaling 17 |
C00003675
|
| 5998 | RGS3, C2PA, RGP3 | regulator of G-protein signaling 3 |
C00003675
|
| 54453 | RIN2, MACS, RASSF4 | Ras and Rab interactor 2 |
C00003675
|
| 79890 | RIN3 | Ras and Rab interactor 3 |
C00003675
|
| 54101 | RIPK4, ANKK2, ANKRD3, DIK, NKRD3, PKK, PPS2, RIP4 | receptor-interacting serine-threonine kinase 4 (EC:2.7.11.1) |
C00003675
|
| 6013 | RLN1, H1, H1RLX, RLXH1, bA12D24.3.1, bA12D24.3.2 | relaxin 1 |
C00003675
|
| 6019 | RLN2, H2, H2-RLX, RLXH2, bA12D24.1.1, bA12D24.1.2 | relaxin 2 |
C00003675
|
| 80010 | RMI1, BLAP75, C9orf76, FAAP75, RP11-346I8.1 | RecQ mediated genome instability 1 |
C00003675
|
| 116028 | RMI2, BLAP18, C16orf75 | RecQ mediated genome instability 2 |
C00003675
|
| 10535 | RNASEH2A, AGS4, JUNB, RNASEHI, RNHIA, RNHL | ribonuclease H2, subunit A (EC:3.1.26.4) |
C00003675
|
| 79621 | RNASEH2B, AGS2, DLEU8 | ribonuclease H2, subunit B |
C00003675
|
| 6041 | RNASEL, PRCA1, RNS4 | ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) (EC:3.1.26.- 3.1.27.-) |
C00003675
|
| 7844 | RNF103, HKF-1, KF-1, KF1, ZFP-103, ZFP103 | ring finger protein 103 |
C00003675
|
| 51444 | RNF138, HSD-4, NARF, STRIN, hNARF | ring finger protein 138, E3 ubiquitin protein ligase |
C00003675
|
| 551328 |
C00003675
|
||
| 55328 | RNLS, C10orf59, RENALASE | renalase, FAD-dependent amine oxidase |
C00003675
|
| 6094 | ROM1, ROM, ROSP1, RP7, TSPAN23 | retinal outer segment membrane protein 1 |
C00003675
|
| 6118 | RPA2, REPA2, RPA32 | replication protein A2, 32kDa |
C00003675
|
| 22934 | RPIA, RPI | ribose 5-phosphate isomerase A (EC:5.3.1.6) |
C00003675
|
| 644937 |
C00003675
|
||
| 6223 | RPS19, DBA, DBA1, S19 | ribosomal protein S19 |
C00003675
|
| 6194 | RPS6, S6 | ribosomal protein S6 |
C00003675
|
| 6195 | RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1) |
C00003675
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00003675
|
| 6198 | RPS6KB1, PS6K, S6K, S6K-beta-1, S6K1, STK14A, p70_S6KA, p70(S6K)-alpha, p70-S6K, p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 (EC:2.7.11.1) |
C00003675
|
| 6237 | RRAS | related RAS viral (r-ras) oncogene homolog |
C00003675
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00003675
|
| 219790 | RTKN2, PLEKHK1, bA531F24.1 | rhotekin 2 |
C00003675
|
| 6253 | RTN2, NSP2, NSPL1, NSPLI, SPG12 | reticulon 2 |
C00003675
|
| 25914 | RTTN | rotatin |
C00003675
|
| 861 | RUNX1, AML1, AML1-EVI-1, AMLCR1, CBFA2, EVI-1, PEBP2aB | runt-related transcription factor 1 |
C00003675
|
| 6281 | S100A10, 42C, ANX2L, ANX2LG, CAL1L, CLP11, Ca[1], GP11, P11, p10 | S100 calcium binding protein A10 |
C00003675
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00003675
|
| 6288 | SAA1, PIG4, SAA, SAA2, TP53I4 | serum amyloid A1 |
C00003675
|
| 6289 | SAA2 | serum amyloid A2 |
C00003675
|
| 148418 | SAMD13, RP11-376N17.1 | sterile alpha motif domain containing 13 |
C00003675
|
| 54809 | SAMD9, C7orf5, DRIF1, NFTC, OEF1, OEF2 | sterile alpha motif domain containing 9 |
C00003675
|
| 163786 | SASS6, SAS-6, SAS6 | spindle assembly 6 homolog (C. elegans) |
C00003675
|
| 66234 |
C00003675
|
||
| 51435 | SCARA3, APC7, CSR, CSR1, MSLR1, MSRL1 | scavenger receptor class A, member 3 |
C00003675
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00003675
|
| 132320 | SCLT1, CAP-1A, CAP1A, hCAP-1A | sodium channel and clathrin linker 1 |
C00003675
|
| 10389 | SCML2 | sex comb on midleg-like 2 (Drosophila) |
C00003675
|
| 6324 | SCN1B, ATFB13, BRGDA5, GEFSP1 | sodium channel, voltage-gated, type I, beta subunit |
C00003675
|
| 6337 | SCNN1A, BESC2, ENaCa, ENaCalpha, SCNEA, SCNN1 | sodium channel, non-voltage-gated 1 alpha subunit |
C00003675
|
| 57758 | SCUBE2, CEGB1, CEGF1, CEGP1 | signal peptide, CUB domain, EGF-like 2 |
C00003675
|
| 27111 | SDCBP2, SITAC, SITAC18, ST-2 | syndecan binding protein (syntenin) 2 |
C00003675
|
| 221935 | SDK1 | sidekick cell adhesion molecule 1 |
C00003675
|
| 6397 | SEC14L1, PRELID4A, SEC14L | SEC14-like 1 (S. cerevisiae) |
C00003675
|
| 23541 | SEC14L2, C22orf6, SPF, TAP, TAP1 | SEC14-like 2 (S. cerevisiae) |
C00003675
|
| 6398 | SECTM1, K12 | secreted and transmembrane 1 |
C00003675
|
| 94009 | SERHL, BK126B4.1, BK126B4.2, HS126B42, dJ222E13.1 | serine hydrolase-like |
C00003675
|
| 347735 | SERINC2, PRO0899, TDE2, TDE2L | serine incorporator 2 |
C00003675
|
| 12 | SERPINA3, AACT, ACT, GIG25 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
C00003675
|
| 5271 | SERPINB8, CAP2, PI8 | serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
C00003675
|
| 5274 | SERPINI1, PI12, neuroserpin | serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
C00003675
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00003675
|
| 6446 | SGK1, SGK | serum/glucocorticoid regulated kinase 1 (EC:2.7.11.1) |
C00003675
|
| 151648 | SGOL1, NY-BR-85, SGO, Sgo1 | shugoshin-like 1 (S. pombe) |
C00003675
|
| 151246 | SGOL2, SGO2, TRIPIN | shugoshin-like 2 (S. pombe) |
C00003675
|
| 63898 | SH2D4A, PPP1R38, SH2A | SH2 domain containing 4A |
C00003675
|
| 6451 | SH3BGRL, SH3BGR | SH3 domain binding glutamic acid-rich protein like |
C00003675
|
| 79729 | SH3D21, C1orf113 | SH3 domain containing 21 |
C00003675
|
| 51100 | SH3GLB1, Bif-1, PPP1R70, dJ612B15.2 | SH3-domain GRB2-like endophilin B1 |
C00003675
|
| 54436 | SH3TC1 | SH3 domain and tetratricopeptide repeats 1 |
C00003675
|
| 26751 | SH3YL1, RAY | SH3 and SYLF domain containing 1 |
C00003675
|
| 6462 | SHBG, ABP, SBP, TEBG | sex hormone-binding globulin |
C00003675
|
| 25759 | SHC2, SCK, SHCB, SLI | SHC (Src homology 2 domain containing) transforming protein 2 |
C00003675
|
| 79801 | SHCBP1, PAL | SHC SH2-domain binding protein 1 |
C00003675
|
| 149345 | SHISA4, C1orf40, TMEM58 | shisa family member 4 |
C00003675
|
| 357 | SHROOM2, APXL, HSAPXL | shroom family member 2 |
C00003675
|
| 6476 | SI | sucrase-isomaltase (alpha-glucosidase) (EC:3.2.1.10 3.2.1.48) |
C00003675
|
| 57568 | SIPA1L2, SPAL2 | signal-induced proliferation-associated 1 like 2 |
C00003675
|
| 100528646 |
C00003675
|
||
| 220134 | SKA1, C18orf24 | spindle and kinetochore associated complex subunit 1 |
C00003675
|
| 348235 | SKA2, FAM33A | spindle and kinetochore associated complex subunit 2 |
C00003675
|
| 221150 | SKA3, C13orf3, RAMA1 | spindle and kinetochore associated complex subunit 3 |
C00003675
|
| 6502 | SKP2, FBL1, FBXL1, FLB1, p45 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
C00003675
|
| 84174 | SLA2, C20orf156, MARS, SLAP-2, SLAP2 | Src-like-adaptor 2 |
C00003675
|
| 84068 | SLC10A7, C4orf13, P7 | solute carrier family 10, member 7 |
C00003675
|
| 6565 | SLC15A2, PEPT2 | solute carrier family 15 (oligopeptide transporter), member 2 |
C00003675
|
| 151473 | SLC16A14, MCT14 | solute carrier family 16, member 14 |
C00003675
|
| 9121 | SLC16A5, MCT5, MCT6 | solute carrier family 16 (monocarboxylate transporter), member 5 |
C00003675
|
| 26503 | SLC17A5, AST, ISSD, NSD, SD, SIALIN, SIASD, SLD | solute carrier family 17 (acidic sugar transporter), member 5 |
C00003675
|
| 116843 | SLC18B1, C6orf192, dJ55C23.6 | solute carrier family 18, subfamily B, member 1 |
C00003675
|
| 5002 | SLC22A18, BWR1A, BWSCR1A, HET, IMPT1, ITM, ORCTL2, SLC22A1L, TSSC5, p45-BWR1A | solute carrier family 22, member 18 |
C00003675
|
| 60386 | SLC25A19, DNC, MCPHA, MUP1, THMD3, THMD4, TPC | solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
C00003675
|
| 788 | SLC25A20, CAC, CACT | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
C00003675
|
| 291 | SLC25A4, 1, AAC1, ANT, ANT_1, ANT1, MTDPS12, PEO2, PEO3, T1 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
C00003675
|
| 1811 | SLC26A3, CLD, DRA | solute carrier family 26 (anion exchanger), member 3 |
C00003675
|
| 376497 | SLC27A1, ACSVL5, FATP, FATP1 | solute carrier family 27 (fatty acid transporter), member 1 |
C00003675
|
| 2030 | SLC29A1, ENT1 | solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
C00003675
|
| 144195 | SLC2A14, GLUT14, SLC2A3P3 | solute carrier family 2 (facilitated glucose transporter), member 14 |
C00003675
|
| 6515 | SLC2A3, GLUT3 | solute carrier family 2 (facilitated glucose transporter), member 3 |
C00003675
|
| 6517 | SLC2A4, GLUT4 | solute carrier family 2 (facilitated glucose transporter), member 4 |
C00003675
|
| 1318 | SLC31A2, COPT2, CTR2, hCTR2 | solute carrier family 31 (copper transporter), member 2 |
C00003675
|
| 201266 | SLC39A11, C17orf26, ZIP11 | solute carrier family 39, member 11 |
C00003675
|
| 254428 | SLC41A1, MgtE | solute carrier family 41 (magnesium transporter), member 1 |
C00003675
|
| 84102 | SLC41A2, SLC41A1-L1 | solute carrier family 41 (magnesium transporter), member 2 |
C00003675
|
| 8501 | SLC43A1, LAT3, PB39, POV1, R00504 | solute carrier family 43 (amino acid system L transporter), member 1 |
C00003675
|
| 23446 | SLC44A1, CD92, CDW92, CHTL1, CTL1, RP11-287A8.1 | solute carrier family 44 (choline transporter), member 1 |
C00003675
|
| 80736 | SLC44A4, C6orf29, CTL4, NG22 | solute carrier family 44, member 4 |
C00003675
|
| 51151 | SLC45A2, 1A1, AIM1, MATP, OCA4, SHEP5 | solute carrier family 45, member 2 |
C00003675
|
| 83959 | SLC4A11, BTR1, CDPD1, CHED2, NABC1, dJ794I6.2 | solute carrier family 4, sodium borate transporter, member 11 |
C00003675
|
| 6531 | SLC6A3, DAT, DAT1, PKDYS | solute carrier family 6 (neurotransmitter transporter), member 3 |
C00003675
|
| 6542 | SLC7A2, ATRC2, CAT2, HCAT2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
C00003675
|
| 22865 | SLITRK3 | SLIT and NTRK-like family, member 3 |
C00003675
|
| 57228 | SMAGP, hSMAGP | small cell adhesion glycoprotein |
C00003675
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00003675
|
| 10051 | SMC4, CAP-C, CAPC, SMC-4, SMC4L1, hCAP-C | structural maintenance of chromosomes 4 |
C00003675
|
| 201895 | SMIM14, C4orf34 | small integral membrane protein 14 |
C00003675
|
| 54498 | SMOX, C20orf16, PAO, PAO-1, PAOH1, SMO | spermine oxidase (EC:1.5.3.16) |
C00003675
|
| 6591 | SNAI2, SLUG, SLUGH1, SNAIL2, WS2D | snail family zinc finger 2 |
C00003675
|
| 333929 | SNAI3, SMUC, SNAIL3, ZNF293, Zfp293 | snail family zinc finger 3 |
C00003675
|
| 6616 | SNAP25, RIC-4, RIC4, SEC9, SNAP, SNAP-25, bA416N4.2, dJ1068F16.2 | synaptosomal-associated protein, 25kDa |
C00003675
|
| 6617 | SNAPC1, PTFgamma, SNAP43 | small nuclear RNA activating complex, polypeptide 1, 43kDa |
C00003675
|
| 6623 | SNCG, BCSG1, SR | synuclein, gamma (breast cancer-specific protein 1) |
C00003675
|
| 85028 | SNHG12, C1orf79, LINC00100, NCRNA00100, RP4-669K10.5 | small nucleolar RNA host gene 12 (non-protein coding) |
C00003675
|
| 83891 | SNX25, SBBI31 | sorting nexin 25 |
C00003675
|
| 257364 | SNX33, SH3PX3, SH3PXD3C, SNX30 | sorting nexin 33 |
C00003675
|
| 8835 | SOCS2, CIS2, Cish2, SOCS-2, SSI-2, SSI2, STATI2 | suppressor of cytokine signaling 2 |
C00003675
|
| 6272 | SORT1, Gp95, LDLCQ6, NT3 | sortilin 1 |
C00003675
|
| 6662 | SOX9, CMD1, CMPD1, SRA1 | SRY (sex determining region Y)-box 9 |
C00003675
|
| 93349 | SP140L | SP140 nuclear body protein-like |
C00003675
|
| 6671 | SP4, HF1B, SPR-1 | Sp4 transcription factor |
C00003675
|
| 6674 | SPAG1, CILD28, HSD-3.8, SP75, TPIS | sperm associated antigen 1 |
C00003675
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00003675
|
| 166378 | SPATA5, AFG2, SPAF | spermatogenesis associated 5 |
C00003675
|
| 147841 | SPC24, SPBC24 | SPC24, NDC80 kinetochore complex component |
C00003675
|
| 57405 | SPC25, SPBC25, hSpc25 | SPC25, NDC80 kinetochore complex component |
C00003675
|
| 54908 | SPDL1, CCDC99 | spindle apparatus coiled-coil protein 1 |
C00003675
|
| 92521 | SPECC1, CYTSB, HCMOGT-1, HCMOGT1, NSP | sperm antigen with calponin homology and coiled-coil domains 1 |
C00003675
|
| 139886 | SPIN4 | spindlin family, member 4 |
C00003675
|
| 6715 | SRD5A1, S5AR_1 | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) (EC:1.3.1.22) |
C00003675
|
| 222183 | SRRM3 | serine/arginine repetitive matrix 3 |
C00003675
|
| 6757 | SSX2, CT5.2, CT5.2A, HD21, HOM-MEL-40, SSX | synovial sarcoma, X breakpoint 2 |
C00003675
|
| 3156 | HMGCR, LDLCQ3 | 3-hydroxy-3-methylglutaryl-CoA reductase (EC:1.1.1.34) |
C00047082
|
| 55808 | ST6GALNAC1, HSY11339, SIAT7A, ST6GalNAcI, STYI | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC:2.4.99.3) |
C00003675
|
| 7982 | ST7, ETS7q, FAM4A, FAM4A1, HELG, RAY1, SEN4, TSG7 | suppression of tumorigenicity 7 |
C00003675
|
| 55620 | STAP2, BKS | signal transducing adaptor family member 2 |
C00003675
|
| 6770 | STAR, STARD1 | steroidogenic acute regulatory protein |
C00003675
|
| 134429 | STARD4 | StAR-related lipid transfer (START) domain containing 4 |
C00003675
|
| 6776 | STAT5A, MGF, STAT5 | signal transducer and activator of transcription 5A |
C00003675
|
| 79689 | STEAP4, STAMP2, TIARP, TNFAIP9 | STEAP family member 4 |
C00003675
|
| 6491 | STIL, MCPH7, SIL | SCL/TAL1 interrupting locus |
C00003675
|
| 9262 | STK17B, DRAK2 | serine/threonine kinase 17b (EC:2.7.11.1) |
C00003675
|
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00003675
|
| 55959 | SULF2, HSULF-2 | sulfatase 2 |
C00003675
|
| 20888 |
C00003675
|
||
| 6839 | SUV39H1, H3-K9-HMTase_1, KMT1A, MG44, SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00003675
|
| 79723 | SUV39H2, KMT1B | suppressor of variegation 3-9 homolog 2 (Drosophila) (EC:2.1.1.43) |
C00003675
|
| 9143 | SYNGR3 | synaptogyrin 3 |
C00003675
|
| 79933 | SYNPO2L | synaptopodin 2-like |
C00003675
|
| 10460 | TACC3, ERIC-1, ERIC1 | transforming, acidic coiled-coil containing protein 3 |
C00003675
|
| 85461 | TANC1, ROLSB, TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
C00003675
|
| 23216 | TBC1D1, TBC, TBC1 | TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
C00003675
|
| 55357 | TBC1D2, PARIS-1, PARIS1, TBC1D2A | TBC1 domain family, member 2 |
C00003675
|
| 11138 | TBC1D8, AD3, HBLP1, TBC1D8A, VRP | TBC1 domain family, member 8 (with GRAM domain) |
C00003675
|
| 6913 | TBX15, TBX14 | T-box 15 |
C00003675
|
| 6916 | TBXAS1, BDPLT14, CYP5, CYP5A1, GHOSAL, THAS, TS, TXAS, TXS | thromboxane A synthase 1 (platelet) (EC:5.3.99.5) |
C00003675
|
| 6941 | TCF19, SC1, TCF-19 | transcription factor 19 |
C00003675
|
| 6949 | TCOF1, MFD1, TCS1, treacle | Treacher Collins-Franceschetti syndrome 1 |
C00003675
|
| 7004 | TEAD4, EFTR-2, RTEF1, TCF13L1, TEF-3, TEF3, TEFR-1, hRTEF-1B | TEA domain family member 4 |
C00003675
|
| 7015 | TERT, CMM9, DKCA2, DKCB4, EST2, PFBMFT1, TCS1, TP2, TRT, hEST2, hTRT | telomerase reverse transcriptase (EC:2.7.7.49) |
C00003675
|
| 93081 | TEX30, C13orf27 | testis expressed 30 |
C00003675
|
| 7023 | TFAP4, AP-4, bHLHc41 | transcription factor AP-4 (activating enhancer binding protein 4) |
C00003675
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00003675
|
| 7035 | TFPI, EPI, LACI, TFI, TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
C00003675
|
| 7037 | TFRC, CD71, T9, TFR, TFR1, TR, TRFR, p90 | transferrin receptor |
C00003675
|
| 7038 | TG, AITD3, TGN | thyroglobulin |
C00003675
|
| 7041 | TGFB1I1, ARA55, HIC-5, HIC5, TSC-5 | transforming growth factor beta 1 induced transcript 1 |
C00003675
|
| 7042 | TGFB2, LDS4, TGF-beta2 | transforming growth factor, beta 2 |
C00003675
|
| 7043 | TGFB3, ARVD, ARVD1, TGF-beta3 | transforming growth factor, beta 3 |
C00003675
|
| 7048 | TGFBR2, AAT3, FAA3, LDS1B, LDS2B, MFS2, RIIC, TAAD2, TGFR-2, TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) (EC:2.7.11.30) |
C00003675
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00003675
|
| 7053 | TGM3, TGE | transglutaminase 3 (EC:2.3.2.13) |
C00003675
|
| 7047 | TGM4, TGP, hTGP | transglutaminase 4 (EC:2.3.2.13) |
C00003675
|
| 9984 | THOC1, HPR1, P84, P84N5 | THO complex 1 |
C00003675
|
| 90381 | TICRR, C15orf42, SLD3, Treslin | TOPBP1-interacting checkpoint and replication regulator |
C00003675
|
| 29090 | TIMM21, C18orf55, TIM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) |
C00003675
|
| 25976 | TIPARP, ARTD14, PARP7, pART14 | TCDD-inducible poly(ADP-ribose) polymerase (EC:2.4.2.30) |
C00003675
|
| 27134 | TJP3, ZO-3, ZO3 | tight junction protein 3 |
C00003675
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00003675
|
| 7098 | TLR3, CD283, IIAE2 | toll-like receptor 3 |
C00003675
|
| 4071 | TM4SF1, H-L6, L6, M3S1, TAAL6 | transmembrane 4 L six family member 1 |
C00003675
|
| 147798 | TMC4 | transmembrane channel-like 4 |
C00003675
|
| 11322 | TMC6, EV1, EVER1, EVIN1, LAK-4P | transmembrane channel-like 6 |
C00003675
|
| 57458 | TMCC3 | transmembrane and coiled-coil domain family 3 |
C00003675
|
| 55002 | TMCO3, C13orf11 | transmembrane and coiled-coil domains 3 |
C00003675
|
| 55273 | TMEM100 | transmembrane protein 100 |
C00003675
|
| 284114 | TMEM102, CBAP | transmembrane protein 102 |
C00003675
|
| 54664 | TMEM106B | transmembrane protein 106B |
C00003675
|
| 84314 | TMEM107, GRVS638, PRO1268 | transmembrane protein 107 |
C00003675
|
| 23306 | TMEM194A, TMEM194 | transmembrane protein 194A |
C00003675
|
| 199953 | TMEM201, NET5 | transmembrane protein 201 |
C00003675
|
| 382245 |
C00003675
|
||
| 140738 | TMEM37, PR, PR1 | transmembrane protein 37 |
C00003675
|
| 148534 | TMEM56 | transmembrane protein 56 |
C00003675
|
| 84283 | TMEM79 | transmembrane protein 79 |
C00003675
|
| 84910 | TMEM87B | transmembrane protein 87B |
C00003675
|
| 7112 | TMPO, CMD1T, LAP2, LEMD4, PRO0868, TP | thymopoietin |
C00003675
|
| 28983 | TMPRSS11E, DESC1, TMPRSS11E2 | transmembrane protease, serine 11E (EC:3.4.21.-) |
C00003675
|
| 7113 | TMPRSS2, PP9284, PRSS10 | transmembrane protease, serine 2 (EC:3.4.21.-) |
C00003675
|
| 11013 | TMSB15A, TMSB15, TMSB15B, TMSL8, TMSNB, Tb15, TbNB | thymosin beta 15a |
C00003675
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00003675
|
| 7128 | TNFAIP3, A20, OTUD7C, TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 (EC:3.4.19.12) |
C00003675
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00003675
|
| 8741 | TNFSF13, APRIL, CD256, TALL-2, TALL2, TRDL-1, ZTNF2 | tumor necrosis factor (ligand) superfamily, member 13 |
C00003675
|
| 9966 | TNFSF15, TL1, TL1A, VEGI, VEGI192A | tumor necrosis factor (ligand) superfamily, member 15 |
C00003675
|
| 4796 | TONSL, IKBR, NFKBIL2 | tonsoku-like, DNA repair protein |
C00003675
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00003675
|
| 11073 | TOPBP1, TOP2BP1 | topoisomerase (DNA) II binding protein 1 |
C00003675
|
| 26092 | TOR1AIP1, LAP1, LAP1B | torsin A interacting protein 1 |
C00003675
|
| 163590 | TOR1AIP2, IFRG15, LULL1, NET9, RP11-12M5.5 | torsin A interacting protein 2 |
C00003675
|
| 27324 | TOX3, CAGF9, TNRC9 | TOX high mobility group box family member 3 |
C00003675
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00003675
|
| 58476 | TP53INP2, C20orf110, DOR, PINH, dJ1181N3.1 | tumor protein p53 inducible nuclear protein 2 |
C00003675
|
| 7162 | TPBG, 5T4, 5T4AG, M6P1 | trophoblast glycoprotein |
C00003675
|
| 7164 | TPD52L1, D53, hD53 | tumor protein D52-like 1 |
C00003675
|
| 22974 | TPX2, C20orf1, C20orf2, DIL-2, DIL2, FLS353, GD:C20orf1, HCA519, HCTP4, REPP86, p100 | TPX2, microtubule-associated |
C00003675
|
| 10293 | TRAIP, RNF206, TRIP | TRAF interacting protein |
C00003675
|
| 9881 | TRANK1, LBA1 | tetratricopeptide repeat and ankyrin repeat containing 1 |
C00003675
|
| 11277 | TREX1, AGS1, CRV, DRN3, HERNS | three prime repair exonuclease 1 (EC:3.1.11.2) |
C00003675
|
| 10206 | TRIM13, CAR, DLEU5, LEU5, RFP2, RNF77 | tripartite motif containing 13 |
C00003675
|
| 10475 | TRIM38, RNF15, RORET | tripartite motif containing 38 |
C00003675
|
| 79097 | TRIM48, RNF101 | tripartite motif containing 48 |
C00003675
|
| 81844 | TRIM56, RNF109 | tripartite motif containing 56 |
C00003675
|
| 286827 | TRIM59, MRF1, RNF104, TRIM57, TSBF1 | tripartite motif containing 59 |
C00003675
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00003675
|
| 10024 | TROAP, TASTIN | trophinin associated protein |
C00003675
|
| 55503 | TRPV6, ABP/ZF, CAT1, CATL, ECAC2, HSA277909, LP6728, ZFAB | transient receptor potential cation channel, subfamily V, member 6 |
C00003675
|
| 7248 | TSC1, LAM, TSC | tuberous sclerosis 1 |
C00003675
|
| 7249 | TSC2, LAM, TSC4 | tuberous sclerosis 2 |
C00003675
|
| 25987 | TSKU, E2IG4, LRRC54, TSK | tsukushi, small leucine rich proteoglycan |
C00003675
|
| 6302 | TSPAN31, SAS | tetraspanin 31 |
C00003675
|
| 22996 | TTC39A, C1orf34, DEME-6 | tetratricopeptide repeat domain 39A |
C00003675
|
| 125488 | TTC39C, C18orf17, HsT2697 | tetratricopeptide repeat domain 39C |
C00003675
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00003675
|
| 7275 | TUB, rd5 | tubby bipartite transcription factor |
C00003675
|
| 406303 |
C00003675
|
||
| 113457 | TUBA3D, H2-ALPHA | tubulin, alpha 3d |
C00003675
|
| 84617 | TUBB6, HsT1601, TUBB-5 | tubulin, beta 6 class V |
C00003675
|
| 1890 | TYMP, ECGF, ECGF1, MEDPS1, MNGIE, MTDPS1, PDECGF, TP, hPD-ECGF | thymidine phosphorylase (EC:2.4.2.4) |
C00003675
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00003675
|
| 11065 | UBE2C, UBCH10, dJ447F3.2 | ubiquitin-conjugating enzyme E2C (EC:6.3.2.19) |
C00003675
|
| 29089 | UBE2T, PIG50 | ubiquitin-conjugating enzyme E2T (putative) (EC:6.3.2.19) |
C00003675
|
| 90025 | UBE3D, C6orf157, H10BH, UBE2CBP, YJR141W | ubiquitin protein ligase E3D (EC:6.3.2.-) |
C00003675
|
| 55148 | UBR7, C14orf130 | ubiquitin protein ligase E3 component n-recognin 7 (putative) |
C00003675
|
| 80019 | UBTD1 | ubiquitin domain containing 1 |
C00003675
|
| 7371 | UCK2, TSA903, UK, UMPK | uridine-cytidine kinase 2 (EC:2.7.1.48) |
C00003675
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00003675
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00003675
|
| 7367 | UGT2B17, BMND12, UDPGT2B17 | UDP glucuronosyltransferase 2 family, polypeptide B17 (EC:2.4.1.17) |
C00003675
|
| 7364 | UGT2B7, UDPGT_2B9, UDPGT2B7, UDPGTH2, UGT2B9 | UDP glucuronosyltransferase 2 family, polypeptide B7 (EC:2.4.1.17) |
C00003675
|
| 29128 | UHRF1, ICBP90, Np95, RNF106, hNP95, hUHRF1 | ubiquitin-like with PHD and ring finger domains 1 (EC:6.3.2.19) |
C00003675
|
| 7374 | UNG, DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 | uracil-DNA glycosylase (EC:3.2.2.27) |
C00003675
|
| 9816 | URB2, KIAA0133, NET10, NPA2 | URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
C00003675
|
| 7398 | USP1, UBP | ubiquitin specific peptidase 1 (EC:3.4.19.12) |
C00003675
|
| 10791 | VAMP5 | vesicle-associated membrane protein 5 |
C00003675
|
| 7431 | VIM, CTRCT30 | vimentin |
C00003675
|
| 23230 | VPS13A, CHAC, CHOREIN | vacuolar protein sorting 13 homolog A (S. cerevisiae) |
C00003675
|
| 7443 | VRK1, PCH1, PCH1A | vaccinia related kinase 1 (EC:2.7.11.1) |
C00003675
|
| 128434 | VSTM2L, C20orf102, dJ1118M15.2 | V-set and transmembrane domain containing 2 like |
C00003675
|
| 4013 | VWA5A, BCSC-1, BCSC1, LOH11CR2A | von Willebrand factor A domain containing 5A |
C00003675
|
| 54838 | WBP1L, C10orf26, OPA1L, OPAL1 | WW domain binding protein 1-like |
C00003675
|
| 115825 | WDFY2, PROF, RP11-147H23.1, WDF2, ZFYVE22 | WD repeat and FYVE domain containing 2 |
C00003675
|
| 11169 | WDHD1, AND-1, CHTF4, CTF4 | WD repeat and HMG-box DNA binding protein 1 |
C00003675
|
| 89891 | WDR34, DIC5, FAP133, bA216B9.3 | WD repeat domain 34 |
C00003675
|
| 284403 | WDR62, C19orf14, MCPH2 | WD repeat domain 62 |
C00003675
|
| 79968 | WDR76, CDW14 | WD repeat domain 76 |
C00003675
|
| 7468 | WHSC1, MMSET, NSD2, REIIBP, TRX5, WHS | Wolf-Hirschhorn syndrome candidate 1 (EC:2.1.1.43) |
C00003675
|
| 55062 | WIPI1, ATG18, ATG18A, WIPI49 | WD repeat domain, phosphoinositide interacting 1 |
C00003675
|
| 7477 | WNT7B | wingless-type MMTV integration site family, member 7B |
C00003675
|
| 7483 | WNT9A, WNT14 | wingless-type MMTV integration site family, member 9A |
C00003675
|
| 55135 | WRAP53, DKCB3, TCAB1, WDR79 | WD repeat containing, antisense to TP53 |
C00003675
|
| 26118 | WSB1, SWIP1, WSB-1 | WD repeat and SOCS box containing 1 |
C00003675
|
| 389610 | XKR5, HARL2754, UNQ2754, XRG5A, XRG5BM | XK, Kell blood group complex subunit-related family, member 5 |
C00003675
|
| 7516 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 |
C00003675
|
| 7517 | XRCC3, CMM6 | X-ray repair complementing defective repair in Chinese hamster cells 3 |
C00003675
|
| 91419 | XRCC6BP1, KUB3 | XRCC6 binding protein 1 (EC:3.4.24.-) |
C00003675
|
| 51776 | ZAK, AZK, MLK7, MLT, MLTK, MRK, mlklak, pk | sterile alpha motif and leucine zipper containing kinase AZK (EC:2.7.11.25) |
C00003675
|
| 7538 | ZFP36, G0S24, GOS24, NUP475, RNF162A, TIS11, TTP, zfp-36 | ZFP36 ring finger protein |
C00003675
|
| 7718 | ZNF165, CT53, LD65, ZSCAN7 | zinc finger protein 165 |
C00003675
|
| 388569 | ZNF324B | zinc finger protein 324B |
C00003675
|
| 195828 | ZNF367, AFF29, CDC14B, ZFF29 | zinc finger protein 367 |
C00003675
|
| 58499 | ZNF462, Zfp462 | zinc finger protein 462 |
C00003675
|
| 348327 | ZNF530 | zinc finger protein 530 |
C00003675
|
| 90233 | ZNF551 | zinc finger protein 551 |
C00003675
|
| 57116 | ZNF695, RP11-551G24.1, SBZF3 | zinc finger protein 695 |
C00003675
|
| 255403 | ZNF718 | zinc finger protein 718 |
C00003675
|
| 55055 | ZWILCH, KNTC1AP, hZwilch | zwilch kinetochore protein |
C00003675
|
| 11130 | ZWINT, HZwint-1, KNTC2AP, ZWINT1 | ZW10 interacting kinetochore protein |
C00003675
|
| 7791 | ZYX, ESP-2, HED-2 | zyxin |
C00003675
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (103)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #100100 | Abdominal muscles, absence of, with urinary tract abnormality and cryptorchidism |
P20309
|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #103780 | Alcohol dependence |
P08172
P14416 P31645 |
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #614373 | Amyotrophic lateral sclerosis 16, juvenile; als16 |
Q99720
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #300908 | Anemia, nonspherocytic hemolytic, due to g6pd deficiency |
P11413
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #602025 | Body mass index quantitative trait locus 9; bmiq9 |
P41968
|
| %606641 | Body mass index; bmi |
P37231
|
| #612560 | Bone mineral density quantitative trait locus 12; bmnd12 |
O75795
|
| #300615 | Brunner syndrome |
P21397
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #302960 | Chondrodysplasia punctata 2, x-linked dominant; cdpx2 |
Q15125
|
| #611489 | Corticosteroid-binding globulin deficiency |
P08185
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #162800 | Cyclic neutropenia |
P08246
|
| #612522 | Diabetes mellitus, insulin-dependent, 22; iddm22 |
P51681
|
| #607208 | Dravet syndrome |
P35498
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #613721 | Epileptic encephalopathy, early infantile, 11; eiee11 |
Q99250
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613659 | Gastric cancer |
P04626
|
| #137215 | Gastric cancer, hereditary diffuse; hdgc |
P04626
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #604403 | Generalized epilepsy with febrile seizures plus, type 2; gefsp2 |
P35498
|
| #231095 | Ghosal hematodiaphyseal dysplasia; ghdd |
P24557
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137800 | Glioma susceptibility 1; glm1 |
P04626
P37231 |
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #609423 | Human immunodeficiency virus type 1, susceptibility to |
P41597
P51681 |
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237450 | Hyperbilirubinemia, rotor type; hblrr |
Q9NPD5
Q9Y6L6 |
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #211980 | Lung cancer |
P00533
P04626 |
| #608516 | Major depressive disorder; mdd |
P08172
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| %300852 | Mental retardation, x-linked 88; mrx88 |
P50052
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #609634 | Migraine, familial hemiplegic, 3; fhm3 |
P35498
|
| #126200 | Multiple sclerosis, susceptibility to; ms |
P08575
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #159900 | Myoclonic dystonia |
P14416
|
| #202700 | Neutropenia, severe congenital, 1, autosomal dominant; scn1 |
P08246
|
| #601665 | Obesity |
P32245
P37231 |
| #164230 | Obsessive-compulsive disorder; ocd |
P31645
|
| #604715 | Orthostatic intolerance |
P23975
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #167000 | Ovarian cancer |
P04626
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #613135 | Parkinsonism-dystonia, infantile; pkdys |
Q01959
|
| #172700 | Pick disease of brain |
P10636
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #607745 | Seizures, benign familial infantile, 3; bfis3 |
Q99250
|
| #608971 | Severe combined immunodeficiency, autosomal recessive, t cell-negative, b cell-positive, nk cell-positive |
P08575
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #190300 | Tremor, hereditary essential, 1; etm1 |
P35462
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #610379 | West nile virus, susceptibility to |
P51681
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (75)
| KEGG | name | UniProt |
|---|---|---|
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04626 (related) |
| H00022 | Bladder cancer |
P00533
(related)
P04626 (related) |
| H00028 | Choriocarcinoma |
P00533
(related)
P03956 (related) P04626 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P04626 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04626 (related) P06401 (marker) Q92731 (marker) |
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
P11511 (related) |
| H00019 | Pancreatic cancer |
P04626
(related)
|
| H00027 | Ovarian cancer |
P04626
(related)
|
| H00031 | Breast cancer |
P04626
(related)
P04626 (marker) |
| H00046 | Cholangiocarcinoma |
P04626
(related)
P35354 (related) |
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H01163 | Corticosteroid-binding globulin (CBG) deficiency |
P08185
(related)
|
| H00100 | Neutropenic disorders |
P08246
(related)
|
| H00091 | T-B+Severe combined immunodeficiencies (SCIDs) |
P08575
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
Q13148 (related) |
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00101 | Other phagocyte defects |
P11413
(related)
|
| H00668 | Anemia due to disorders of glutathione metabolism |
P11413
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00025 | Penile cancer |
P14780
(related)
P35354 (related) |
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
Q92887 (related) |
| H01031 | Orthostatic intolerance (OI) |
P23975
(related)
|
| H00490 | Diaphyseal dysplasia with anemia (Ghosal) |
P24557
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00775 | Familial or sporadic hemiplegic migraine |
P35498
(related)
|
| H00783 | Febrile seizures |
P35498
(related)
|
| H00032 | Thyroid cancer |
P37231
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
P50052
(related)
Q99714 (related) |
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|
| H01194 | X-linked chondrodysplasia punctata |
Q15125
(related)
|
| H00606 | Early infantile epileptic encephalopathy |
Q99250
(related)
|
| H00806 | Benign familial neonatal and infantile epilepsies |
Q99250
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q9NUW8
(related)
|
Diseases related to CTD interactions
93 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000230 | Adenocarcinoma |
C00003675
|
| D000647 | Amnesia |
C00003675
|
| D000787 | Angina Pectoris |
C00003675
|
| D000987 | Antisocial Personality Disorder |
C00003675
|
| D001008 | Anxiety Disorders |
C00003675
|
| D050197 | Atherosclerosis |
C00003675
|
| D001724 | Birth Weight |
C00003675
|
| D002545 | Brain Ischemia |
C00003675
|
| D001943 | Breast Neoplasms |
C00003675
|
| D002056 | Burns |
C00003675
|
| D002318 | Cardiovascular Diseases |
C00003675
|
| D002375 | Catalepsy |
C00003675
|
| D002561 | Cerebrovascular Disorders |
C00003675
|
| D002659 | Child Development Disorders, Pervasive |
C00003675
|
| D019970 | Cocaine-Related Disorders |
C00003675
|
| D003327 | Coronary Disease |
C00003675
|
| D003456 | Cryptorchidism |
C00003675
|
| D003866 | Depressive Disorder |
C00003675
|
| D003865 | Depressive Disorder, Major |
C00003675
|
| D004195 | Disease Models, Animal |
C00003675
|
| D012734 | Disorders of Sex Development |
C00003675
|
| D004409 | Dyskinesia, Drug-Induced |
C00003675
|
| D019263 | Dysthymic Disorder |
C00003675
|
| D004831 | Epilepsies, Myoclonic |
C00003675
|
| D007172 | Erectile Dysfunction |
C00003675
|
| D005221 | Fatigue |
C00003675
|
| D005317 | Fetal Growth Retardation |
C00003675
|
| D018222 | Fibromatosis, Aggressive |
C00003675
|
| D005355 | Fibrosis |
C00003675
|
| D005391 | Firesetting Behavior |
C00003675
|
| D005832 | Genital Diseases, Male |
C00003675
|
| D005909 | Glioblastoma |
C00003675
|
| D006331 | Heart Diseases |
C00003675
|
| D006333 | Heart Failure |
C00003675
|
| D006628 | Hirsutism |
C00003675
|
| D019247 | HIV Wasting Syndrome |
C00003675
|
| D019584 | Hot Flashes |
C00003675
|
| D006869 | Hydronephrosis |
C00003675
|
| D006930 | Hyperalgesia |
C00003675
|
| D017588 | Hyperandrogenism |
C00003675
|
| D006948 | Hyperkinesis |
C00003675
|
| D006973 | Hypertension |
C00003675
|
| D006984 | Hypertrophy |
C00003675
|
| D007006 | Hypogonadism |
C00003675
|
| D007018 | Hypopituitarism |
C00003675
|
| D007021 | Hypospadias |
C00003675
|
| D007174 | Impulse Control Disorders |
C00003675
|
| D007248 | Infertility, Male |
C00003675
|
| D007333 | Insulin Resistance |
C00003675
|
| D007680 | Kidney Neoplasms |
C00003675
|
| D007713 | Klinefelter Syndrome |
C00003675
|
| D007859 | Learning Disorders |
C00003675
|
| D007984 | Leydig Cell Tumor |
C00003675
|
| D020347 | Lithiasis |
C00003675
|
| D008113 | Liver Neoplasms |
C00003675
|
| D008228 | Lymphoma, Non-Hodgkin |
C00003675
|
| D008288 | Malaria |
C00003675
|
| D015674 | Mammary Neoplasms, Animal |
C00003675
|
| D008325 | Mammary Neoplasms, Experimental |
C00003675
|
| D008546 | Melanoma, Experimental |
C00003675
|
| D001523 | Mental Disorders |
C00003675
|
| D009133 | Muscular Atrophy |
C00003675
|
| D017202 | Myocardial Ischemia |
C00003675
|
| D009205 | Myocarditis |
C00003675
|
| D009362 | Neoplasm Metastasis |
C00003675
|
| D009369 | Neoplasms |
C00003675
|
| D009395 | Nephritis, Interstitial |
C00003675
|
| D009422 | Nervous System Diseases |
C00003675
|
| D020258 | Neurotoxicity Syndromes |
C00003675
|
| D010146 | Pain |
C00003675
|
| D010300 | Parkinson Disease |
C00003675
|
| D011085 | Polycystic Ovary Syndrome |
C00003675
|
| D011086 | Polycythemia |
C00003675
|
| D011297 | Prenatal Exposure Delayed Effects |
C00003675
|
| D015175 | Prolactinoma |
C00003675
|
| D011470 | Prostatic Hyperplasia |
C00003675
|
| D019048 | Prostatic Intraepithelial Neoplasia |
C00003675
|
| D011471 | Prostatic Neoplasms |
C00003675
|
| D011472 | Prostatitis |
C00003675
|
| D015427 | Reperfusion Injury |
C00003675
|
| D012735 | Sexual Dysfunction, Physiological |
C00003675
|
| D020018 | Sexual Dysfunctions, Psychological |
C00003675
|
| D012771 | Shock, Hemorrhagic |
C00003675
|
| D012774 | Shock, Traumatic |
C00003675
|
| D020181 | Sleep Apnea, Obstructive |
C00003675
|
| D013226 | Status Epilepticus |
C00003675
|
| D013375 | Substance Withdrawal Syndrome |
C00003675
|
| D013927 | Thrombosis |
C00003675
|
| D014646 | Varicocele |
C00003675
|
| D061205 | Vascular Calcification |
C00003675
|
| D014770 | Virilism |
C00003675
|
| D014832 | Voice Disorders |
C00003675
|
| D015430 | Weight Gain |
C00003675
|