Agelas dispar
Species
KNApSAcK Entry
| Organism name | Agelas dispar |
|---|---|
| Genus | |
| Family | |
| Kingdom |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Agelas dispar |
|---|---|
| Linked NCBI taxonomy ID | 295252 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Agelasidae |
|---|---|
| ID | 85771 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Metazoa |
|---|---|
| ID | 33208 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (11)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00003643
|
Ajugasterone C
|
CHEMBL477336
|
1 / 1 / 0 | No. 58 | No. 11 |

|
||
|
C00049800
|
Keramadine
|
CHEMBL1162276
|
No. 2659 |

|
||||
|
C00028214
|
Dispacamide C
|
CHEMBL73258
|
No. 3889 |

|
||||
|
C00049784
|
Clathramide D
/ (+)-Clathramide D |
CHEMBL465780
CHEMBL464126 |
No. 4756 |

|
||||
|
C00049783
|
Clathramide C
|
CHEMBL465780
CHEMBL464126 |
No. 4756 |

|
||||
|
C00049806
|
Longamide B
|
CHEMBL465779
|
No. 5211 |

|
||||
|
C00039037
|
Dispyrin
|
CHEMBL493091
|
4 / 2 / 2 | No. 5970 |

|
|||
|
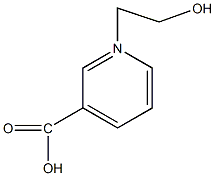
C00050207
|
Pyridinebetaine A
|
No. 7888 |
|
|||||
|
C00043577
|
Homarine
|
C010981
|
No. 8014 |
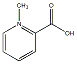
|
||||
|
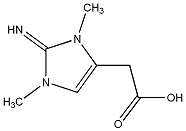
C00027844
|
Aminozooanemonin
|
CHEMBL442935
|
No. 8872 |
|
||||
|
C00050208
|
Pyridinebetaine B
|
No. 8941 |

|
Human Protein / Gene in interactions
5 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00039037 | 2 / 2 |
| Q9Y5N1 | Histamine H3 receptor | Histamine receptor | C00039037 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00003643 | 1 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00039037 | 0 / 0 |
| P25100 | Alpha-1D adrenergic receptor | Adrenergic receptor | C00039037 | 0 / 0 |