E-zyme 2 for prediction of enzymatic reactions
About E-zyme 2
E-zyme 2 is a server for identifying an orphan enzyme using chemical structures of substrate-product pair (reactant pair). The methods is based on a search for similar reactant pairs in RPAIR database and provides ortholog groups (KO and OC) that possibly mediate the given reactant pair.
Usage
- Input a pair of compound in MDL mol format, or following compound identifiers.
- KEGG COMPOUND ID (e.g. C00077)
- KEGG DRUG ID (e.g. D08032)
- PubChem CID (e.g. CID6262)
- PubChem SID (e.g. SID3377)
- NIKKAJI ID (e.g. J9.177D)
- KNApSAcK ID (e.g. C00001384)
- Click the "view structures" button.
- Click the "compute" button if compound images are shown.
Output
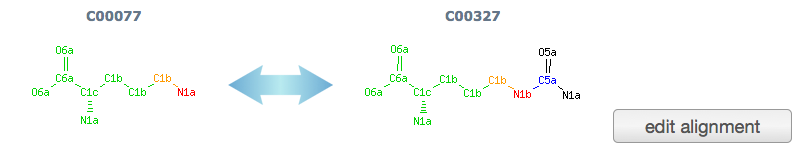
alignment: The atom alignment of the query reactant pair by SIMCOMP program. Atom color shows matched (green, yellow and red) and unmatched region (black and blue). (see SIMCOMP help, RDM patterns)
When the auto-alignment of atom was wrong, user can edit alignment manually from "edit alignment" button.
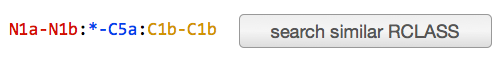
RDM pattern: The representation of chemical changes occurring during the query reaction. User can search reactant pairs with similar pattern from "search similar RCLASS" button. (see RPAIR, RCLASS)
prediction: The list of ortholog groups (KO and OC) that possibly mediate the query reactant pair. IDs colored in orange are prokaryote spesific ortholog group, IDs colored in green are eukaryote specific group, and gray IDs are not spesific group. Prediction scores are based on the similarity of alignment structures of reference reactant pairs.
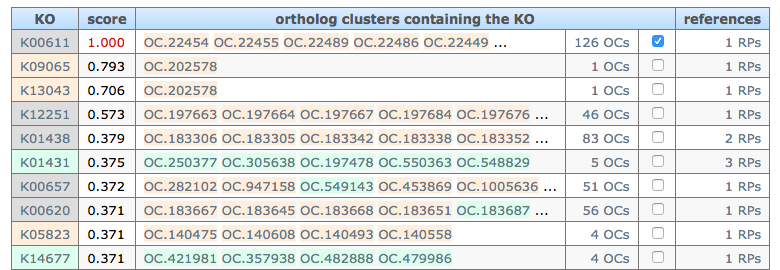
User can search genes of a specific organism, or ortholog groups (OCs) that contain specific taxon, from predicted OCs whose checkbox is checked in the table.
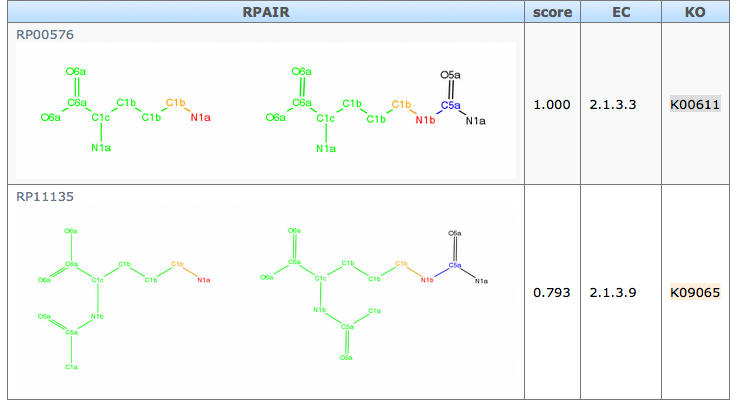
all reference pairs: The list of referenced reactant pair for orpahn enzyme prediction. Scores are based on the similarity of alignment structure.
Similarity between reactant pairs
To define the similarity between reactant pairs, each reactant pair was converted to the substructure profile, an integer vector that describes the aligned substructure and its frequency. Each aligned substructure is represented in joining of KEGG atom, hyphen (that shows alignment of atoms), and coron (that shows bond in the substrate or the product).
example of substructure profile
| Aligned substructure | Frequency |
|---|---|
| C1b-C1b:N1a-N1b | 1 |
| N1a-N1b:*-C5a | 1 |
| C1b-C1b:C1b-C1b:N1a-N1b | 1 |
| C1b-C1b:N1a-N1b:*-C5a | 1 |
| ... | ... |
The TANIMOTO coefficient between the profile of the reactant pairs is used as a measure of similarity.
Reference
- Moriya, Y., Yamada, T., Okuda, S., Nakagawa, Z., Kotera, M., Tokimatsu, T., Kanehisa, M., Goto, S.; Identification of Enzyme Genes Using Chemical Structure Alignments of Substrate-Product Pairs. J. Chem. Inf. Model. (2016). [pubmed]