Query form
Reference pathway:
PathPred provides the RDM pattern libraries for two reference pathways; the biodegradation of xenobiotics in bacteria and the biosynthesis of secondary metabolites in plants, which utilizes different characteristic subsets of the RDM patterns. The users can choose one of the reference pathway according to their purpose.
- RPAIR list of xenobiotics biodegradation
- RPAIR list of biosynthesis of secondary metabolites
Enter initial, final compound:
Input a query compound in the MDL mol file format, in the SMILES representation, or by the KEGG compound identifier. When the reference pathway is xenobiotics biodegradation, the query should be the compound to undergo biodegradation. And when the reference pathway is biosynthesis of secondary metabolites, the query should be the end product of biosynthesis.
Optionally, a final compound in the biodegradation or a initial compound in the biosynthesis can be also specified, if the compound is already known (bi-directional prediction).
Options:
E-mail Adrress: The URL to access the results will be sent to this address after the prediction is completed.
Simcomp Threshold: Set the threshold of global similarity searching by SIMCOMP program (SIMCOMP help). The default threshold is set as 0.3. When the value is near 1, the prediction proceeds by using compounds with high similarity to query as reference.
Prediction cycle: Set the number of prediction cycles of the "cycle B" that is described below. You should increase the value when the prediction doesn't reache the last compound of biodegradation or biosynthesis.
< Example 1 > (Tetrachrolobenzene biodegradation (a few minutes))
< Example 2 > (Fraxidin biosynthesis (a few minutes))
< Example 3 > (Geniodelpin biosynthesis (30 minutes))
Result
Predicted pathway tree:
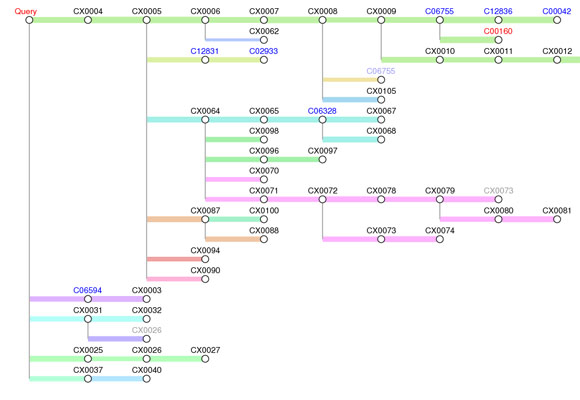
The predicted pathway tree consists compounds (node) and reactions (edge). The blue-colored C numbers are compounds in the KEGG pathway and the black-colored CX numbers are compounds not stored in KEGG. A greyed-out node means that there is the same compound elsewhere in the tree. When the pathway reaches to the final compound in bi-directional prediction, the compound is highlighted in red. The consecutive reactions in same color show that the reference reactions are consecutive in KEGG pathway. The thickness of the line shows the plausibility of the predicted reaction based on the reaction score.

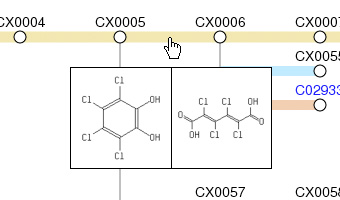 Structure images popup when the mouse is moved over each object if JavaScript is enebled in the web browser.
Structure images popup when the mouse is moved over each object if JavaScript is enebled in the web browser.
Predicted pathway:
In each predicted pathway, the reaction scores and the pathway scores from the query to the compound are shown.
Predicted reaction:
predicted reaction
reference rpairs
In each predicted reaction, the reference RPAIR images colored by RDM pattern are shown. And the related KEGG REACTION and KEGG Orthology (KO) are linked.
Method
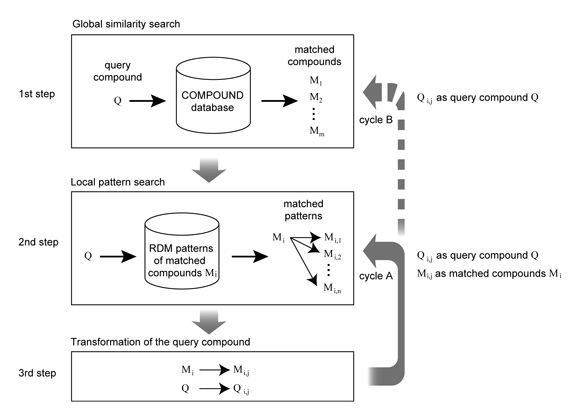
The first step of our method is a global similarity search of a query compound Q against the KEGG COMPOUND database by the SIMCOMP program, a maximal common subgraph search program. The second step is a local similarity search against the RDM patterns of matched compounds Mi, to select the matched patterns Mi --> Mi,j that are applicable to the query compound. The third step is transformation of the query compound Q to generate compounds Qi,j by following the matched patterns. The second and third steps are repeated (cycle A) as long as the generated compounds Qi,j and Mi,j can be used in the same way as the query compound Q and the matched compounds Mi. If not, the transformed compounds Qi,j are used as next queries for the global chemical structure search against the COMPOUND database (cycle B). This prediction cycle B may be repeated for a given number of times. The PathPred accepts bi-directional prediction when the user inputs the starting and final compounds. Then the prediction cycle is also performed from the final compound, which would limit the possible search space, and is repeated until the predicted pathway is connected to that from the initial query compound or the prediction cycle is repeated for a given number of times.
Reference
Moriya Y, Shigemizu D, Hattori M, Tokimatsu T, Kotera M, Goto S, Kanehisa M.; PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 38, Web Server issue (2010). [pubmed] [NAR]
Feedback
Please use the feedback page to send your comments or questions to PathPred.
GenomeNet Feedback Form
|

