ViPTree
: the Viral Proteomic Tree server
version 4.0
ViPTree
: the Viral Proteomic Tree server
version 4.0
About
The ViPTree server generates a "proteomic tree" of viral genome sequences based on genome-wide sequence similarities computed by tBLASTx.
The original proteomic tree concept (i.e., "the Phage Proteomic Tree”) was developed by Rohwer and Edwards, 2002.
A proteomic tree is a dendrogram that reveals global genomic similarity relationships between tens, hundreds, and thousands of viruses.
It has been shown that viral groups identified in a proteomic tree well correspond to established viral taxonomies.
The proteomic tree approach is effective to investigate genomes of newly sequenced viruses as well as those identified in metagenomes.
VipTree also offers an excellent alignment visualization useful for comparative genomics of viruses.
You may also try DiGAlign, which extends VipTree's alignment visualization features that can be used for all nucleotide sequences, including non-viral genomes.
Workflow
- Select nucleic acid types: dsDNA / dsDNA-RT / ssDNA / dsRNA / ssRNA / ssRNA-RT
- Select host categories: prokaryotic / eukaryotic / any host
- Browse proteomic trees and genomic alignments between reference viruses
- Download figures and tables
Generate a proteomic tree of your viruses
- Upload your viral sequences and choose reference viruses to generate a proteomic tree
- Browse the proteomic tree
- Regenerate a focused proteomic tree only with viruses selected from the entire proteomic tree for interpretation / presentation
- Browse dotplots / genomic alignments between your viruses and reference viruses
- Browse genomic similarity scores and gene annotation
- Download figures and tables
Example
Here is an exmaple of the session main page (an entrance page after the computation).
Snapshots
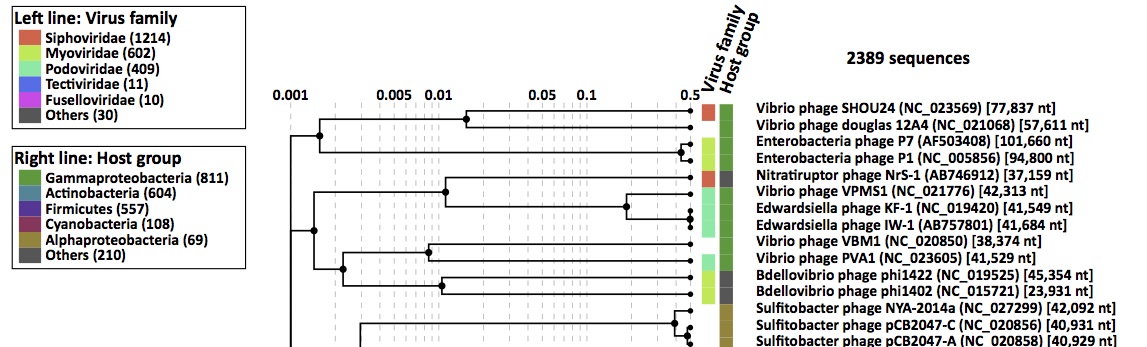
Rectangular proteomic tree
Previous
Next
- Circular proteomic tree: trees of prokaryotic dsDNA viruses (left) and eukaryotic ssDNA viruses (right), colored by virus families and host taxonomic groups.
- Rectangular proteomic tree: a part of a dsDNA viruses tree, shown with virus names. Each of the inner nodes links to a genomic alignment of genomes that are included in its subtree.
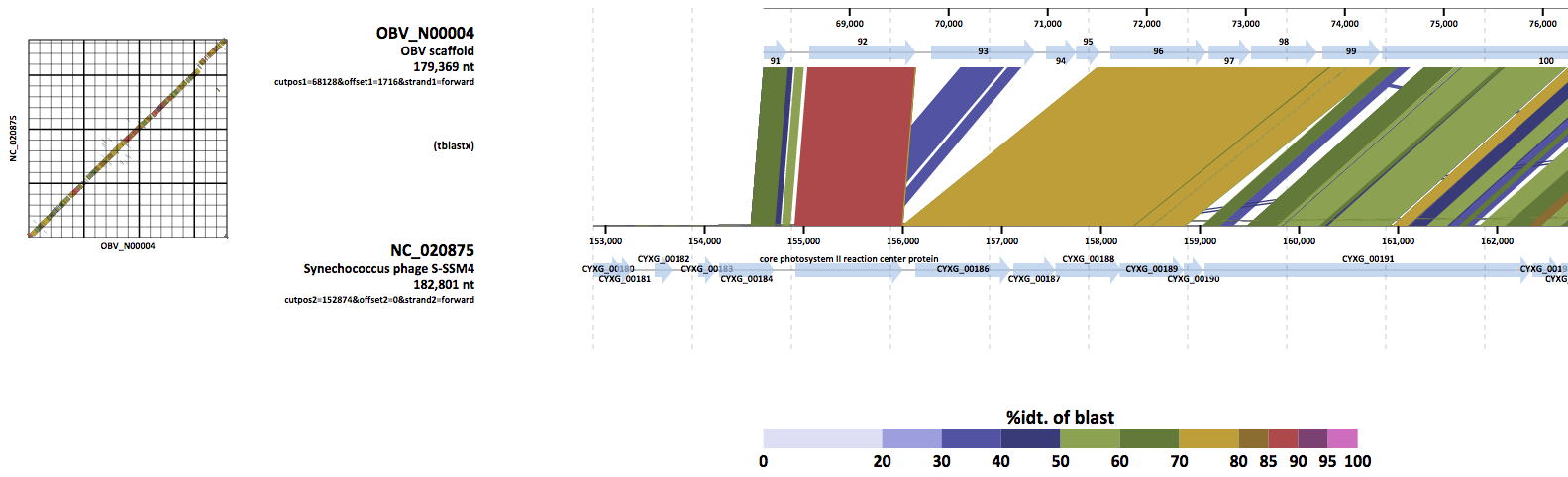
- Genomic alignmnet of metagenomic genome and Synechococcus phage S-SSM4.
Version history
- 2023-11-17 version 4.0
- Major update! Various functions are newly implemented, as listed below.
- Gene prediction is now performed by Prodigal and any coding table is selectable if it is compatible with Prodigal.
- In the alignment viewer, threshold for tBLASTx/BLASTn could be set.
- In the alignment viewer, a mouseover popup is implemented that shows details of genes and tBLASTx/BLASTn hits.
- From this version, License is set as 'CC BY-NC-SA 4.0'
- Many bug fixes, including the gene table upload function.
- Document is updated.
- Version of Virus-Host DB: RefSeq release 220
- 2023-08-11 version 3.7
- Version of Virus-Host DB: RefSeq release 219
- 2023-06-02 version 3.6
- Version of Virus-Host DB: RefSeq release 218
- 2023-04-03 version 3.5
- Version of Virus-Host DB: RefSeq release 217
- 2023-02-13 version 3.4
- Version of Virus-Host DB: RefSeq release 216
- 2022-12-9 version 3.3
- Version of Virus-Host DB: RefSeq release 215
- 2022-11-11 version 3.2
- Version of Virus-Host DB: RefSeq release 214
- 2022-06-20 version 3.1
- Version of Virus-Host DB: RefSeq release 212
- 2022-04-18 version 3.0
- Version of Virus-Host DB: RefSeq release 211
- The upload function is updated.
- New options of the alignment is available.
- 2022-03-14 version 2.1
- Version of Virus-Host DB: RefSeq release 210
- 2021-12-13 version 2.0
- Version of Virus-Host DB: RefSeq release 209
- "Only query" mode was launched.
- 2021-10-04 version 1.9.1
- Version of Virus-Host DB: RefSeq release 207
- Selectable nucleic acid types is changed.(old version was dsDNA/ssDNA/dsRNA/ssRNA/retroviruses/satellite viruses and virophages)
- 2019-04-22 version 1.9
- Version of Virus-Host DB: RefSeq release 93
- 2019-02-12 version 1.8
- Version of Virus-Host DB: RefSeq release 92
- 2018-12-25 version 1.7
- Version of Virus-Host DB: RefSeq release 91
- 2018-11-05 version 1.6
- Version of Virus-Host DB: RefSeq release 90
- 2018-08-24 version 1.5
- Version of Virus-Host DB: RefSeq release 89
- 2018-06-22 version 1.4
- Version of Virus-Host DB: RefSeq release 88
- 2018-04-24 version 1.3
- Version of Virus-Host DB: RefSeq release 87
- 2018-03-27 version 1.2
- Version of Virus-Host DB: RefSeq release 86
- New feature: gene annotaion method can be selected from prokaryotic, eukaryotic, and user defined genes.
- Improved usability: the number of the max upload sequences is expanded to 50.
- Improved usability: long sequence ID (up to 30 characters) is acceptable. Sequence IDs can include dots and hyphens.
- Improved usability: Colors in alignment view can be customized.
- 2017-02-08 version 1.1
- Version of Virus-Host DB: RefSeq release 76
- New feature: "related" proteomic tree is available in user sessions
- 2017-01-04 version 1.0
- Version of Virus-Host DB: RefSeq release 76
- Initial release of ViPTree
Code availability
We developed
ViPTreeGen, a command line tool for viral proteomic tree generation.
Reference virus data
Reference viral sequences and taxonomic information are available at
Virus-Host DB.
Related work
We published a study of over thousand marine EVGs (environmental viral genomes). In the study, a prototype of the ViPTree server was developed and applied to the analysis.
Supplemental page for the study (including proteomic tree viewer and alignment viewer) can be browsed.
Citation
If you use results (data / figures) genereted by ViPTree in your research, please cite:
Following papers are related in methodological aspects to the ViPTree server.
- Nishimura, Y. et al. Environmental viral genomes shed new light on virus–host interactions in the ocean.
mSphere, 2, e00359-16, doi:10.1128/mSphere.00359-16 (2017) [PubMed]
- Bhunchoth, Y. et al. Two asian jumbo phages, ϕRSL2 and ϕRSF1, infect Ralstonia solanacearum and show common features of ϕKZ-related phages.
Virology, 494, 56-66, doi:10.1016/j.virol.2016.03.028 (2016).
Feedback
We appreciate bug reports, comments, and suggestions. The form is
here.