KCF-S cluster No. 243 (27 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| asterids | 50 |
Cumulative family count
| class name | count |
|---|---|
| Oleaceae | 48 |
| Apiaceae | 1 |
| Acanthaceae | 1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (27)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00003093
|
Oleuropein
|
CHEMBL1911053
|
C002769
|
8 / 5 |

|
|
|
C00010761
|
Isoligustroside
|

|
||||
|
C00010766
|
Jasminoside
|

|
||||
|
C00010777
|
Ligustaloside B
|

|
||||
|
C00010778
|
Ligustaloside A
|

|
||||
|
C00010785
|
Ligustroside
|
CHEMBL1086877
|
1 / 1 / 0 |

|
||
|
C00010786
|
10-Hydroxyoleuropein
|

|
||||
|
C00010787
|
10-Acetoxyligustroside
|

|
||||
|
C00010789
|
Oleuroside
|

|
||||
|
C00033126
|
Lucidumoside A
/ (-)-Lucidumoside A |

|
||||
|
C00033127
|
Lucidumoside B
|

|
||||
|
C00033128
|
Lucidumoside C
/ (-)-Lucidumoside C |

|
||||
|
C00033129
|
Lucidumoside D
|

|
||||
|
C00034379
|
2''-Epifraxamoside
|

|
||||
|
C00036342
|
(2''R)-2''-Hydroxyoleuropein
|

|
||||
|
C00036343
|
(2''S)-2''-Hydroxyoleuropein
|

|
||||
|
C00037025
|
Demethylligstroside
/ (-)-Demethylligstroside |

|
||||
|
C00037161
|
Fraxamoside
/ (-)-Fraxamoside |

|
||||
|
C00037335
|
Isooleuropein
|

|
||||
|
C00037421
|
Ligstroside
/ (-)-Ligstroside |
CHEMBL1086877
|
1 / 1 / 0 |

|
||
|
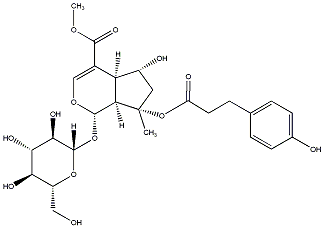
C00037770
|
Saletpangponoside C
/ (-)-Saletpangponoside C |
|
||||
|
C00049064
|
10-Hydroxyligstroside
/ (-)-10-Hydroxyligstroside |

|
||||
|
C00050125
|
Jaslanceoside A
/ (-)-Jaslanceoside A |

|
||||
|
C00050126
|
Jaslanceoside B
/ (-)-Jaslanceoside B |

|
||||
|
C00050127
|
Jaslanceoside C
/ (-)-Jaslanceoside C |

|
||||
|
C00050128
|
Jaslanceoside D
/ (-)-Jaslanceoside D |

|
||||
|
C00050129
|
Jaslanceoside E
/ (-)-Jaslanceoside E |

|
Human Protein / Gene in interactions
1 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P14672 | Solute carrier family 2, facilitated glucose transporter member 4 | Unclassified protein | C00010785 C00037421 | 1 / 0 |
8 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00003093
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00003093
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00003093
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00003093
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00003093
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00003093
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00003093
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00003093
|