KCF-S cluster No. 3405 (3 metabolites)
Corresponding Phytochemical cluster No. 1
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| Liliopsida | 3 |
Cumulative family count
| class name | count |
|---|---|
| Arecaceae | 3 |
KEGG BRITE br08003
Categories (1)
| br08003 Category | # of metabolite |
|---|---|
| Pyridine alkaloids | 1 |
metabolites link (1)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|---|---|
| Pyridine alkaloids | C10129 | C00002020 |
Metabolite list (3)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00002020
|
Arecoline
|
CHEMBL7303
|
D001115
|
15 / 22 / 18 | 64 / 21 |
|
|
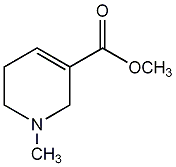
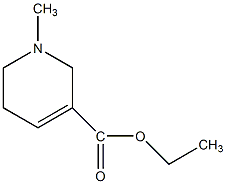
C00044165
|
Ethyl N-methyl-1,2,5,6-tetrahydro-pyridine-3-carboxylate
|
|
||||
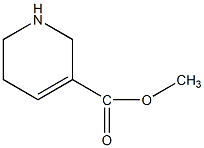
|
C00044191
|
Guvacoline
|
CHEMBL268808
|
C094105
|
1 / 1 / 0 |
|
Human Protein / Gene in interactions
15 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P20309 | Muscarinic acetylcholine receptor M3 | Acetylcholine receptor | C00002020 C00044191 | 1 / 0 |
| P11229 | Muscarinic acetylcholine receptor M1 | Acetylcholine receptor | C00002020 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00002020 | 11 / 10 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00002020 | 3 / 1 |
| P08912 | Muscarinic acetylcholine receptor M5 | Acetylcholine receptor | C00002020 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00002020 | 0 / 1 |
| P08172 | Muscarinic acetylcholine receptor M2 | Acetylcholine receptor | C00002020 | 2 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00002020 | 2 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00002020 | 1 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00002020 | 0 / 0 |
| P08173 | Muscarinic acetylcholine receptor M4 | Acetylcholine receptor | C00002020 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00002020 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002020 | 0 / 1 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00002020 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00002020 | 1 / 4 |
64 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 57491 | AHRR, AHH, AHHR, bHLHe77 | aryl-hydrocarbon receptor repressor |
C00002020
|
| 10327 | AKR1A1, ALDR1, ALR, ARM, DD3 | aldo-keto reductase family 1, member A1 (aldehyde reductase) (EC:1.1.1.2) |
C00002020
|
| 223 | ALDH9A1, ALDH4, ALDH7, ALDH9, E3, TMABADH | aldehyde dehydrogenase 9 family, member A1 (EC:1.2.1.3 1.2.1.19 1.2.1.47) |
C00002020
|
| 328 | APEX1, APE, APE1, APEN, APEX, APX, HAP1, REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 (EC:4.2.99.18) |
C00002020
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00002020
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00002020
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00002020
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00002020
|
| 902 | CCNH, CAK, p34, p37 | cyclin H |
C00002020
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00002020
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00002020
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00002020
|
| 8208 | CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7, MPP7 | chromatin assembly factor 1, subunit B (p60) |
C00002020
|
| 1131 | CHRM3, EGBRS, HM3 | cholinergic receptor, muscarinic 3 |
C00002020
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002020
|
| 56603 | CYP26B1, CYP26A2, P450RAI-2, P450RAI2, RHFCA | cytochrome P450, family 26, subfamily B, polypeptide 1 |
C00002020
|
| 64858 | DCLRE1B, APOLLO, SNM1B, SNMIB | DNA cross-link repair 1B |
C00002020
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00002020
|
| 3301 | DNAJA1, DJ-2, DjA1, HDJ2, HSDJ, HSJ2, HSPF4, NEDD7, hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 |
C00002020
|
| 9521 | EEF1E1, AIMP3, P18 | eukaryotic translation elongation factor 1 epsilon 1 |
C00002020
|
| 2068 | ERCC2, COFS2, EM9, TTD, XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 (EC:3.6.4.12) |
C00002020
|
| 2189 | FANCG, FAG, XRCC9 | Fanconi anemia, complementation group G |
C00002020
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002020
|
| 2937 | GSS, GSHS | glutathione synthetase (EC:6.3.2.3) |
C00002020
|
| 2966 | GTF2H2, BTF2, BTF2P44, T-BTF2P44, TFIIH, p44 | general transcription factor IIH, polypeptide 2, 44kDa |
C00002020
|
| 404672 | GTF2H5, C6orf175, TFB5, TFIIH, TGF2H5, TTD, TTD-A, TTDA, bA120J8.2 | general transcription factor IIH, polypeptide 5 |
C00002020
|
| 51182 | HSPA14, HSP70-4, HSP70L1 | heat shock 70kDa protein 14 |
C00002020
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002020
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002020
|
| 51451 | LCMT1, LCMT, PPMT1 | leucine carboxyl methyltransferase 1 (EC:2.1.1.233) |
C00002020
|
| 4128 | MAOA, MAO-A | monoamine oxidase A (EC:1.4.3.4) |
C00002020
|
| 100302175 | MIR1207, MIRN1207, hsa-mir-1207 | microRNA 1207 |
C00002020
|
| 100188847 | MIR1225, MIRN1225 | microRNA 1225 |
C00002020
|
| 100302142 | MIR1246, MIRN1246, hsa-mir-1246 | microRNA 1246 |
C00002020
|
| 100302233 | MIR1268A, MIR1268, MIRN1268, hsa-mir-1268, hsa-mir-1268a | microRNA 1268a |
C00002020
|
| 100302276 | MIR1290, MIRN1290, hsa-mir-1290 | microRNA 1290 |
C00002020
|
| 100302270 | MIR1305, MIRN1305, hsa-mir-1305 | microRNA 1305 |
C00002020
|
| 406927 | MIR136, MIRN136, miRNA136 | microRNA 136 |
C00002020
|
| 406935 | MIR143, MIRN143 | microRNA 143 |
C00002020
|
| 406937 | MIR145, MIRN145, miR-145, miRNA145 | microRNA 145 |
C00002020
|
| 100302137 | MIR1914, MIRN1914, hsa-mir-1914 | microRNA 1914 |
C00002020
|
| 100302129 | MIR1915, MIRN1915, hsa-mir-1915 | microRNA 1915 |
C00002020
|
| 406978 | MIR199B, MIRN199B, mir-199b | microRNA 199b |
C00002020
|
| 407010 | MIR23A, MIRN23A, hsa-mir-23a, miRNA23A | microRNA 23a |
C00002020
|
| 407015 | MIR26A1, MIR26A, MIRN26A1 | microRNA 26a-1 |
C00002020
|
| 407017 | MIR26B, MIRN26B, hsa-mir-26b, miR-26b | microRNA 26b |
C00002020
|
| 407026 | MIR29C, MIRN29C, miRNA29C | microRNA 29c |
C00002020
|
| 407029 | MIR30A, MIRN30A | microRNA 30a |
C00002020
|
| 693215 | MIR630, MIRN630, hsa-mir-630 | microRNA 630 |
C00002020
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00002020
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00002020
|
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00002020
|
| 4489 | MT1A, MT1, MT1S, MTC | metallothionein 1A |
C00002020
|
| 9027 | NAT8, ATase2, CML1, GLA, Hcml1, TSC501, TSC510 | N-acetyltransferase 8 (GCN5-related, putative) |
C00002020
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00002020
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002020
|
| 5810 | RAD1, HRAD1, REC1 | RAD1 homolog (S. pombe) (EC:3.1.11.2) |
C00002020
|
| 10111 | RAD50, NBSLD, RAD502, hRad50 | RAD50 homolog (S. cerevisiae) |
C00002020
|
| 6283 | S100A12, CAAF1, CAGC, CGRP, ENRAGE, MRP-6, MRP6, p6 | S100 calcium binding protein A12 |
C00002020
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00002020
|
| 7076 | TIMP1, CLGI, EPA, EPO, HCI, TIMP | TIMP metallopeptidase inhibitor 1 |
C00002020
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00002020
|
| 56886 | UGGT1, HUGT1, UGCGL1, UGT1 | UDP-glucose glycoprotein glucosyltransferase 1 |
C00002020
|
| 100126299 | VTRNA2-1, CBL-3, CBL3, MIR886, MIRN886, VTRNA2, hsa-mir-886, hvg-5, nc886 | vault RNA 2-1 |
C00002020
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (22)
| OMIM | preferred title | UniProt |
|---|---|---|
| #100100 | Abdominal muscles, absence of, with urinary tract abnormality and cryptorchidism |
P20309
|
| #103780 | Alcohol dependence |
P08172
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #608516 | Major depressive disorder; mdd |
P08172
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
KEGG DISEASE (18)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
Diseases related to CTD interactions
21 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000647 | Amnesia |
C00002020
|
| D001919 | Bradycardia |
C00002020
|
| D002277 | Carcinoma |
C00002020
|
| D002280 | Carcinoma, Basal Cell |
C00002020
|
| D002375 | Catalepsy |
C00002020
|
| D020820 | Dyskinesias |
C00002020
|
| D006965 | Hyperplasia |
C00002020
|
| D007022 | Hypotension |
C00002020
|
| D007153 | Immunologic Deficiency Syndromes |
C00002020
|
| D009062 | Mouth Neoplasms |
C00002020
|
| D009127 | Muscle Rigidity |
C00002020
|
| D009374 | Neoplasms, Experimental |
C00002020
|
| D019954 | Neurobehavioral Manifestations |
C00002020
|
| D009914 | Oral Submucous Fibrosis |
C00002020
|
| D010212 | Papilloma |
C00002020
|
| D010243 | Paralysis |
C00002020
|
| D020734 | Parkinsonian Disorders |
C00002020
|
| D011470 | Prostatic Hyperplasia |
C00002020
|
| D013226 | Status Epilepticus |
C00002020
|
| D014062 | Tongue Neoplasms |
C00002020
|
| D014202 | Tremor |
C00002020
|