KCF-S cluster No. 3692 (3 metabolites)
Corresponding Phytochemical cluster No. 68
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| rosids | 3 |
| asterids | 2 |
| Liliopsida | 1 |
| eudicotyledons | 1 |
Cumulative family count
| class name | count |
|---|---|
| Brassicaceae | 2 |
| Caprifoliaceae | 1 |
| Sapotaceae | 1 |
| Vitaceae | 1 |
| Arecaceae | 1 |
| Ganodermataceae | 1 |
| Caryophyllaceae | 1 |
KEGG BRITE br08003
Categories (1)
| br08003 Category | # of metabolite |
|---|---|
| Saturated fatty acids | 2 |
metabolites link (2)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|---|---|
| Saturated fatty acids | C00246 | C00001180 |
| Saturated fatty acids | C01585 | C00001218 |
Metabolite list (3)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00001180
|
Butyric acid
|
CHEMBL14227
|
D020148
|
12 / 3 / 2 | 40 / 5 |
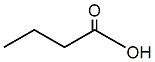
|
|
C00001208
|
Valeric acid
|
CHEMBL268736
|
C038780
|
1 / 7 / 3 |
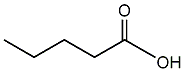
|
|
|
C00001218
|
Caproic acid
/ Hexanoic acid / 1-Hexanoic acid / n-Hexanoic acid |
CHEMBL14184
|
C037652
|
5 / 0 / 0 |
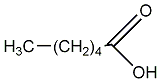
|
Human Protein / Gene in interactions
16 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00001180 C00001218 | 0 / 0 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00001180 C00001218 | 0 / 0 |
| O15379 | Histone deacetylase 3 | Hydrolase | C00001180 | 0 / 0 |
| Q92769 | Histone deacetylase 2 | Hydrolase | C00001180 | 0 / 0 |
| Q9UQL6 | Histone deacetylase 5 | Hydrolase | C00001180 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00001218 | 0 / 0 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00001208 | 7 / 3 |
| Q9UGP5 | DNA polymerase lambda | Enzyme | C00001218 | 0 / 0 |
| Q9BY41 | Histone deacetylase 8 | Hydrolase | C00001180 | 2 / 0 |
| Q7Z4W1 | L-xylulose reductase | Enzyme | C00001180 | 0 / 1 |
| Q13547 | Histone deacetylase 1 | Hydrolase | C00001180 | 0 / 0 |
| P56524 | Histone deacetylase 4 | Hydrolase | C00001180 | 1 / 1 |
| Q8WUI4 | Histone deacetylase 7 | Hydrolase | C00001180 | 0 / 0 |
| Q9UBN7 | Histone deacetylase 6 | Hydrolase | C00001180 | 0 / 0 |
| Q9UKV0 | Histone deacetylase 9 | Hydrolase | C00001180 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00001218 | 0 / 0 |
40 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 5243 | ABCB1, ABC20, CD243, CLCS, GP170, MDR1, P-GP, PGY1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 (EC:3.6.3.44) |
C00001180
|
| 567 | B2M | beta-2-microglobulin |
C00001180
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00001180
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00001180
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00001180
|
| 1081 | CGA, CG-ALPHA, FSHA, GPHA1, GPHa, HCG, LHA, TSHA | glycoprotein hormones, alpha polypeptide |
C00001180
|
| 64581 | CLEC7A, BGR, CANDF4, CLECSF12, DECTIN1 | C-type lectin domain family 7, member A |
C00001180
|
| 2002 | ELK1 | ELK1, member of ETS oncogene family |
C00001180
|
| 8288 | EPX, EPO, EPP, EPX-PEN | eosinophil peroxidase (EC:1.11.1.7) |
C00001180
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00001180
|
| 2623 | GATA1, ERYF1, GATA-1, GF-1, GF1, NF-E1, NFE1, XLANP, XLTDA, XLTT | GATA binding protein 1 (globin transcription factor 1) |
C00001180
|
| 2624 | GATA2, DCML, MONOMAC, NFE1B | GATA binding protein 2 |
C00001180
|
| 2993 | GYPA, CD235a, GPA, GPErik, GPSAT, HGpMiV, HGpMiXI, HGpSta(C), MN, MNS, PAS-2 | glycophorin A (MNS blood group) |
C00001180
|
| 10013 | HDAC6, CPBHM, HD6 | histone deacetylase 6 (EC:3.5.1.98) |
C00001180
|
| 8290 | HIST3H3, H3.4, H3/g, H3FT, H3t | histone cluster 3, H3 |
C00001180
|
| 121504 | HIST4H4, H4/p | histone cluster 4, H4 |
C00001180
|
| 3145 | HMBS, PBG-D, PBGD, PORC, UPS | hydroxymethylbilane synthase (EC:2.5.1.61) |
C00001180
|
| 3606 | IL18, IGIF, IL-18, IL-1g, IL1F4 | interleukin 18 (interferon-gamma-inducing factor) |
C00001180
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00001180
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00001180
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00001180
|
| 10524 | KAT5, ESA1, HTATIP, HTATIP1, PLIP, TIP, TIP60, ZC2HC5, cPLA2 | K(lysine) acetyltransferase 5 (EC:2.3.1.48) |
C00001180
|
| 4254 | KITLG, FPH2, KL-1, Kitl, MGF, SCF, SF, SHEP7 | KIT ligand |
C00001180
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00001180
|
| 4778 | NFE2, NF-E2, p45 | nuclear factor, erythroid 2 |
C00001180
|
| 11315 | PARK7, DJ-1, DJ1 | parkinson protein 7 |
C00001180
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00001180
|
| 5553 | PRG2, BMPG, MBP, MBP1 | proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
C00001180
|
| 5742 | PTGS1, COX1, COX3, PCOX1, PES-1, PGG/HS, PGHS-1, PGHS1, PHS1, PTGHS | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00001180
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00001180
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00001180
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00001180
|
| 6732 | SRPK1, SFRSK1 | SRSF protein kinase 1 (EC:2.7.11.1) |
C00001180
|
| 6733 | SRPK2, SFRSK2 | SRSF protein kinase 2 (EC:2.7.11.1) |
C00001180
|
| 6427 | SRSF2, PR264, SC-35, SC35, SFRS2, SFRS2A, SRp30b | serine/arginine-rich splicing factor 2 |
C00001180
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00001180
|
| 7043 | TGFB3, ARVD, ARVD1, TGF-beta3 | transforming growth factor, beta 3 |
C00001180
|
| 7096 | TLR1, CD281, TIL, TIL._LPRS5, rsc786 | toll-like receptor 1 |
C00001180
|
| 10333 | TLR6, CD286 | toll-like receptor 6 |
C00001180
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00001180
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (10)
| OMIM | preferred title | UniProt |
|---|---|---|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #600430 | Brachydactyly-mental retardation syndrome; bdmr |
P56524
|
| #300882 | Cornelia de lange syndrome 5; cdls5 |
Q9BY41
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #309585 | Wilson-turner x-linked mental retardation syndrome; wts |
Q9BY41
|
KEGG DISEASE (5)
| KEGG | name | UniProt |
|---|---|---|
| H00561 | Brachydacytly-mental retardation syndrome and Smith-Magenis syndrome |
P56524
(related)
|
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H01065 | Pentosuria |
Q7Z4W1
(related)
|