Rubia cordifolia L.
Species
KNApSAcK Entry
| Organism name | Rubia cordifolia L. |
|---|---|
| Genus | Rubia |
| Family | Rubiaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Rubia cordifolia |
|---|---|
| Linked NCBI taxonomy ID | 339321 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Rubiaceae |
|---|---|
| ID | 24966 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Natural Activity
List (17)
| Species | Activity |
|---|---|
| Rubia cordifolia L. | Alexeteric |
| Rubia cordifolia L. | Alterative |
| Rubia cordifolia L. | Analgesic |
| Rubia cordifolia L. | Antiseptic |
| Rubia cordifolia L. | Aperitif |
| Rubia cordifolia L. | Astringent |
| Rubia cordifolia L. | Bitter |
| Rubia cordifolia L. | Delirriant |
| Rubia cordifolia L. | Deobstruent |
| Rubia cordifolia L. | Diuretic |
| Rubia cordifolia L. | Emmenagogue |
| Rubia cordifolia L. | Hypoglycemic |
| Rubia cordifolia L. | Lactagogue |
| Rubia cordifolia L. | Laxative |
| Rubia cordifolia L. | Tonic |
| Rubia cordifolia L. | Vermifuge |
| Rubia cordifolia L. | Vulnerary |
Metabolite list (6)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00019064
|
Oleanolic acid
/ Astrantiagenin C / Virgaureagenin B / 3beta-Hydroxyolean-12-en-28-oic acid |
CHEMBL56615
CHEMBL168 CHEMBL180553 CHEMBL365375 CHEMBL486382 CHEMBL1413646 CHEMBL1436454 |
D009828
|
30 / 8 / 12 | 21 / 15 | No. 13 | No. 51 |
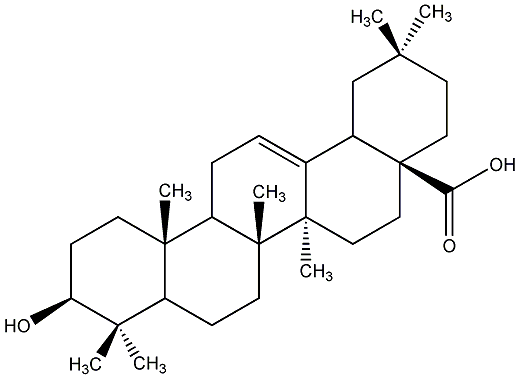
|
|
C00003672
|
Sitosterol
/ beta-sitosterl / (-)-beta-Sitosterol / Stigmast-5-en-3beta-ol |
CHEMBL221542
CHEMBL1398443 CHEMBL1875388 |
17 / 19 / 12 | No. 53 | No. 11 |
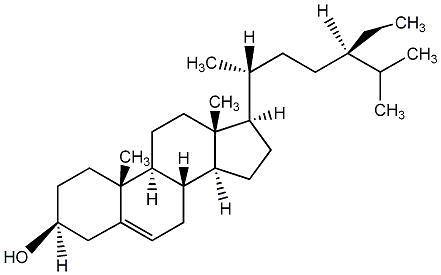
|
||
|
C00033565
|
Acetyloleanolic acid
/ Oleanolic acid acetate / 3-O-Acetyloleanolic acid / (+)-Acetyloleanolic acid / (+)-Oleanolic acid acetate / (+)-3-O-Acetyloleanolic acid / 3beta-Acetoxyolean-12-en-28-oic acid |
CHEMBL486822
CHEMBL494653 CHEMBL1315416 |
C052658
|
1 / 0 / 0 | No. 177 |
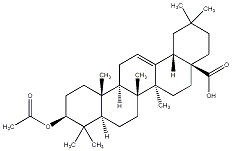
|
||
|
C00029473
|
3,3'-bis(3,4-dihydro-4-hydroxy-6-methoxy-2H-1-benzopyran)
|
No. 3579 |

|
|||||
|
C00030781
|
Mollugin
|
CHEMBL446008
|
C046245
|
9 / 0 | No. 5069 |

|
||
|
C00030333
|
Furomollugin
|
CHEMBL464791
|
No. 8597 |

|
Human Protein / Gene in interactions
44 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00003672 C00019064 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00003672 C00019064 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00019064 C00033565 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00003672 C00019064 | 0 / 0 |
| P00734 | Prothrombin | S1A | C00003672 | 4 / 2 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00019064 | 0 / 0 |
| Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Tyr | C00019064 | 4 / 2 |
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | Tyr | C00019064 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00019064 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00003672 | 2 / 0 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00003672 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00003672 | 3 / 2 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00019064 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00003672 | 1 / 0 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003672 | 0 / 1 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00019064 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00019064 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00019064 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00019064 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00003672 | 1 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00019064 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00019064 | 0 / 3 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00019064 | 2 / 2 |
| P14679 | Tyrosinase | Oxidoreductase | C00003672 | 4 / 2 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00019064 | 0 / 0 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00019064 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00003672 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00019064 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00003672 | 1 / 1 |
| P35228 | Nitric oxide synthase, inducible | Enzyme | C00019064 | 1 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003672 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00003672 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00019064 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00019064 | 0 / 0 |
| P17706 | Tyrosine-protein phosphatase non-receptor type 2 | Tyr | C00019064 | 0 / 1 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | Tyr | C00019064 | 0 / 0 |
| P04054 | Phospholipase A2 | Enzyme | C00019064 | 0 / 0 |
| P80365 | Corticosteroid 11-beta-dehydrogenase isozyme 2 | Enzyme | C00003672 | 1 / 1 |
| P23469 | Receptor-type tyrosine-protein phosphatase epsilon | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00003672 | 1 / 1 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00019064 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00019064 | 1 / 4 |
28 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00019064
C00030781
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00019064
C00030781
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00019064
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00030781
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00030781
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00030781
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00030781
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00030781
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00030781
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00019064
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00019064
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00019064
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00019064
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00019064
|
| 100506742 | CASP12, CASP-12, CASP12P1 | caspase 12 (gene/pseudogene) |
C00030781
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00019064
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00019064
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00019064
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00019064
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00019064
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00019064
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00019064
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00019064
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00019064
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00019064
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00019064
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00019064
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00019064
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (27)
| OMIM | preferred title | UniProt |
|---|---|---|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #218030 | Apparent mineralocorticoid excess; ame |
P80365
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #607785 | Juvenile myelomonocytic leukemia; jmml |
Q06124
|
| #151100 | Leopard syndrome 1 |
Q06124
|
| #611162 | Malaria, susceptibility to |
P35228
|
| #156250 | Metachondromatosis; metcds |
Q06124
|
| #163950 | Noonan syndrome 1; ns1 |
Q06124
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #601367 | Stroke, ischemic |
P00734
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
KEGG DISEASE (24)
| KEGG | name | UniProt |
|---|---|---|
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00038 | Malignant melanoma |
P14679
(marker)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00408 | Type I diabetes mellitus |
P17706
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00017 | Esophageal cancer |
P35228
(related)
P35354 (related) |
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00259 | Apparent mineralocorticoid excess syndrome |
P80365
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q06124
(related)
|
| H01018 | Metachondromatosis |
Q06124
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q9NUW8
(related)
|
Diseases related to CTD interactions
15 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D002252 | Carbon Tetrachloride Poisoning |
C00019064
|
| D056486 | Drug-Induced Liver Injury |
C00019064
|
| D050171 | Dyslipidemias |
C00019064
|
| D018149 | Glucose Intolerance |
C00019064
|
| D006949 | Hyperlipidemias |
C00019064
|
| D007249 | Inflammation |
C00019064
|
| D007674 | Kidney Diseases |
C00019064
|
| D008103 | Liver Cirrhosis |
C00019064
|
| D008106 | Liver Cirrhosis, Experimental |
C00019064
|
| D008107 | Liver Diseases |
C00019064
|
| D017202 | Myocardial Ischemia |
C00019064
|
| D009369 | Neoplasms |
C00019064
|
| D009765 | Obesity |
C00019064
|
| D011041 | Poisoning |
C00019064
|
| D011230 | Precancerous Conditions |
C00019064
|