Brassica spp.
Species
KNApSAcK Entry
| Organism name | Brassica spp. |
|---|---|
| Genus | Brassica |
| Family | Cruciferae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Brassica |
|---|---|
| Linked NCBI taxonomy ID | 3705 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Brassicaceae |
|---|---|
| ID | 3700 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (1)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00003776
|
Lutein
/ (all-E)-Lutein / all-trans-(+)-Xanthophyll / (3R,3'R,6'R)-beta,epsilon-Carotene-3,3'-diol |
CHEMBL173929
CHEMBL172477 CHEMBL1559643 CHEMBL1979448 |
D014975
|
4 / 2 / 3 | 7 / 2 | No. 26 | No. 59 |
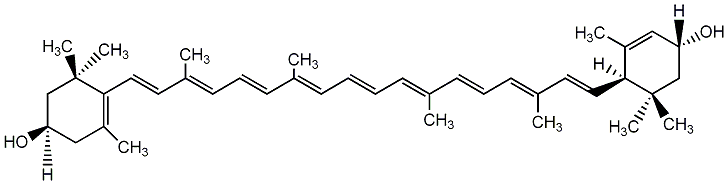
|
Human Protein / Gene in interactions
4 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P11473 | Vitamin D3 receptor | NR1I1 | C00003776 | 2 / 3 |
| O75496 | Geminin | Unclassified protein | C00003776 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00003776 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00003776 | 0 / 0 |
7 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00003776
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00003776
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00003776
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00003776
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00003776
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00003776
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00003776
|