Chlorella vulgaris
Species
KNApSAcK Entry
| Organism name | Chlorella vulgaris |
|---|---|
| Genus | Chlorella |
| Family | Chlorellaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Chlorella vulgaris |
|---|---|
| Linked NCBI taxonomy ID | 3077 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Chlorellaceae |
|---|---|
| ID | 35461 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | Viridiplantae |
|---|---|
| ID | 33090 |
Metabolite list (2)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
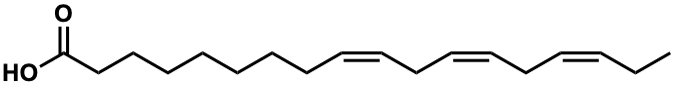
C00007247
|
Linolenic acid
/ alpha-Linolenic acid / (Z,Z,Z)-Octadeca-9,12,15-trienoic acid |
D017962
|
22 / 5 |
|
||||
|
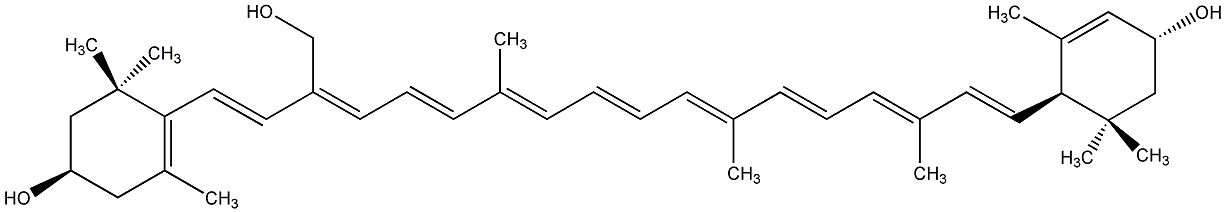
C00022916
|
Loroxanthin
/ (3R,3'R,6'R)-beta,epsilon-Carotene-3,3',19-triol |
No. 26 | No. 59 |
|
Human Protein / Gene in interactions
22 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 8647 | ABCB11, ABC16, BRIC2, BSEP, PFIC-2, PFIC2, PGY4, SPGP | ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
C00007247
|
| 1374 | CPT1A, CPT1, CPT1-L, L-CPT1 | carnitine palmitoyltransferase 1A (liver) (EC:2.3.1.21) |
C00007247
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00007247
|
| 1591 | CYP24A1, CP24, CYP24, HCAI, P450-CC24 | cytochrome P450, family 24, subfamily A, polypeptide 1 (EC:1.14.13.126) |
C00007247
|
| 1557 | CYP2C19, CPCJ, CYP2C, P450C2C, P450IIC19 | cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00007247
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00007247
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00007247
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00007247
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00007247
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00007247
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00007247
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00007247
|
| 3565 | IL4, BCGF-1, BCGF1, BSF-1, BSF1, IL-4 | interleukin 4 |
C00007247
|
| 3827 | KNG1, BDK, BK, KNG | kininogen 1 |
C00007247
|
| 9971 | NR1H4, BAR, FXR, HRR-1, HRR1, RIP14 | nuclear receptor subfamily 1, group H, member 4 |
C00007247
|
| 5465 | PPARA, NR1C1, PPAR, PPARalpha, hPPAR | peroxisome proliferator-activated receptor alpha |
C00007247
|
| 100136658 |
C00007247
|
||
| 5467 | PPARD, FAAR, NR1C2, NUC1, NUCI, NUCII, PPARB | peroxisome proliferator-activated receptor delta |
C00007247
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00007247
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00007247
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00007247
|
| 7421 | VDR, NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
C00007247
|