Streptomyces I-523
Species
KNApSAcK Entry
| Organism name | Streptomyces I-523 |
|---|---|
| Genus | Streptomyces |
| Family | Streptomycetaceae |
| Kingdom | Bacteria |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Streptomyces |
|---|---|
| Linked NCBI taxonomy ID | 1883 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Streptomycetaceae |
|---|---|
| ID | 2062 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Bacteria |
|---|---|
| ID | 2 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (1)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
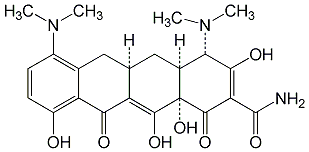
C00018686
|
Vectrin
/ Minocin / Periocline / NSC 141993 / Klinomycin / Minocycline chloride / Minocycline hydrochloride |
CHEMBL1434
CHEMBL1515060 CHEMBL1619602 |
22 / 5 / 5 | No. 5500 |
|
Human Protein / Gene in interactions
22 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00018686 | 1 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00018686 | 0 / 0 |
| Q9Y694 | Solute carrier family 22 member 7 | Unclassified protein | C00018686 | 0 / 0 |
| P02768 | Serum albumin | Secreted protein | C00018686 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00018686 | 0 / 1 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00018686 | 0 / 0 |
| Q8TCC7 | Solute carrier family 22 member 8 | Antiporter | C00018686 | 0 / 0 |
| P55085 | Proteinase-activated receptor 2 | Protease-activated receptor | C00018686 | 0 / 0 |
| Q4U2R8 | Solute carrier family 22 member 6 | Antiporter | C00018686 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00018686 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00018686 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00018686 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00018686 | 0 / 1 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00018686 | 0 / 0 |
| Q9UM07 | Protein-arginine deiminase type-4 | Enzyme | C00018686 | 1 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00018686 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00018686 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00018686 | 1 / 1 |
| Q9NSA0 | Solute carrier family 22 member 11 | Transporter | C00018686 | 0 / 0 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00018686 | 0 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00018686 | 1 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00018686 | 0 / 0 |
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (5)
| OMIM | preferred title | UniProt |
|---|---|---|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #180300 | Rheumatoid arthritis; ra |
Q9UM07
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|