Equisetum debile
Species
KNApSAcK Entry
| Organism name | Equisetum debile |
|---|---|
| Genus | Equisetum |
| Family | Equisetaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Equisetum ramosissimum subsp. debile |
|---|---|
| Linked NCBI taxonomy ID | 232203 |
| Linked level | subspecies |
Family
| Family in NCBI taxonomy | Equisetaceae |
|---|---|
| ID | 3256 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | Euphyllophyta |
|---|---|
| ID | 78536 |
Metabolite list (7)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00005182
|
Kaempferol 3,7-diglucoside
/ Kaempferol 3,7-O-beta-D-diglucopyranoside |
CHEMBL2206209
|
No. 1 | No. 15 |
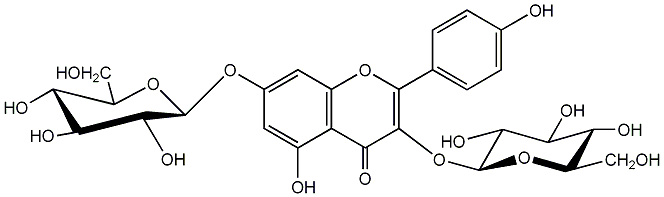
|
|||
|
C00005165
|
Sophoraflavonoloside
/ Kaempferol 3-O-sophoroside / Kaempferol 3-O-beta-sophoroside |
CHEMBL500233
CHEMBL1375795 CHEMBL2032411 |
C064309
|
12 / 2 / 2 | 6 / 4 | No. 1 | No. 15 |
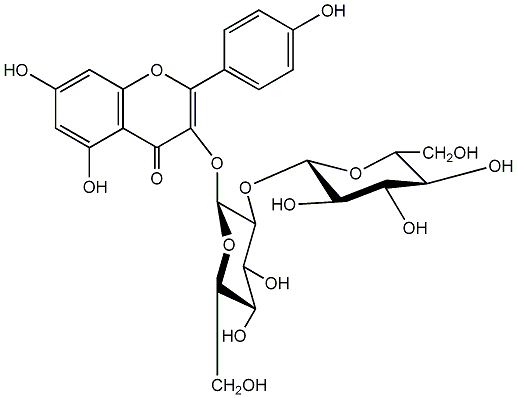
|
|
C00005229
|
(-)-Kaempferol 3-O-sophoroside 7-O-beta-D-glucopyranoside
/ Kaempferol 3-O-sophoroside 7-O-beta-D-glucopyranoside:Kaempferol 3-sophoroside-7-glucoside |
No. 5 | No. 15 |
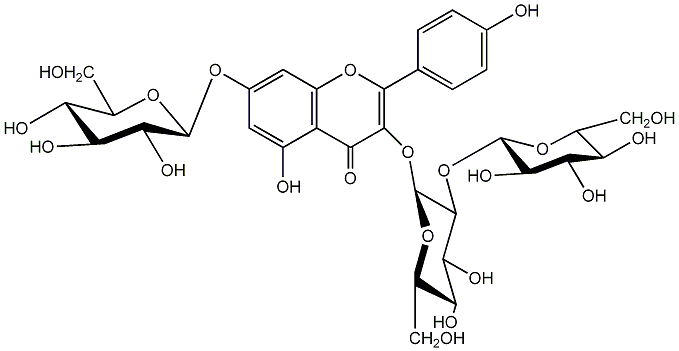
|
||||
|
C00031280
|
Sammangaoside A
/ Staphylionoside H / (-)-Sammangaoside A |
CHEMBL1814430
|
No. 153 |

|
||||
|
C00030700
|
Macarangioside D
|
No. 225 |

|
|||||
|
C00030097
|
Debiloside B
/ Staphylionoside F / (-)-Staphylionoside F |
No. 316 |

|
|||||
|
C00001396
|
L-Tryptophan
|
CHEMBL54976
CHEMBL292303 CHEMBL484901 |
49 / 41 / 39 | No. 1432 | No. 4 |
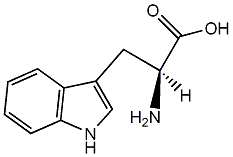
|
Human Protein / Gene in interactions
55 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P39748 | Flap endonuclease 1 | Enzyme | C00001396 C00005165 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00001396 C00005165 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00001396 C00005165 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00001396 C00005165 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00001396 C00005165 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00001396 C00005165 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00001396 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00001396 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00001396 | 0 / 0 |
| Q16773 | Kynurenine--oxoglutarate transaminase 1 | Enzyme | C00001396 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00005165 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00001396 | 11 / 10 |
| Q9Y263 | Phospholipase A-2-activating protein | Unclassified protein | C00001396 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00001396 | 3 / 2 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00001396 | 1 / 2 |
| P02768 | Serum albumin | Secreted protein | C00001396 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00001396 | 0 / 1 |
| P54132 | Bloom syndrome protein | Enzyme | C00001396 | 1 / 2 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00001396 | 0 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00001396 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00001396 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00001396 | 1 / 2 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00001396 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00001396 | 1 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00005165 | 1 / 1 |
| P42858 | Huntingtin | Unclassified protein | C00001396 | 1 / 1 |
| P06746 | DNA polymerase beta | Enzyme | C00005165 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00001396 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00005165 | 1 / 1 |
| P09923 | Intestinal-type alkaline phosphatase | Enzyme | C00001396 | 0 / 0 |
| P05186 | Alkaline phosphatase, tissue-nonspecific isozyme | Enzyme | C00001396 | 3 / 1 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00001396 | 1 / 2 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00001396 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00001396 | 0 / 1 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00001396 | 1 / 1 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00001396 | 5 / 2 |
| P48775 | Tryptophan 2,3-dioxygenase | Enzyme | C00001396 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00001396 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00001396 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00001396 | 0 / 1 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00001396 | 2 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00001396 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00001396 | 4 / 3 |
| Q99549 | M-phase phosphoprotein 8 | Unclassified protein | C00001396 | 0 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00001396 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00001396 | 1 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00001396 | 1 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00005165 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00001396 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00005165 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00001396 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00001396 | 2 / 5 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00001396 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00001396 | 1 / 4 |
| Q05BR4 | SLC16A10 protein | Unclassified protein | C00001396 | 0 / 0 |
6 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00005165
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00005165
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00005165
|
| 7097 | TLR2, CD282, TIL4 | toll-like receptor 2 |
C00005165
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00005165
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00005165
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (43)
| OMIM | preferred title | UniProt |
|---|---|---|
| #210900 | Bloom syndrome; blm |
P54132
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #143100 | Huntington disease; hd |
P42858
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #146300 | Hypophosphatasia, adult |
P05186
|
| #241510 | Hypophosphatasia, childhood |
P05186
|
| #241500 | Hypophosphatasia, infantile |
P05186
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (41)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00213 | Hypophosphatasia |
P05186
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00059 | Huntington's disease (HD) |
P42858
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00208 | Hyperbilirubinemia |
Q92887
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|