Ligusticum scoticum
Species
KNApSAcK Entry
| Organism name | Ligusticum scoticum |
|---|---|
| Genus | Ligusticum |
| Family | Apiaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Ligusticum scoticum |
|---|---|
| Linked NCBI taxonomy ID | 49554 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Apiaceae |
|---|---|
| ID | 4037 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (2)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
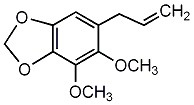
C00002737
|
Dillapiole
|
CHEMBL470874
|
C059115
|
1 / 0 / 0 | No. 1917 | No. 6 |
|
|
|
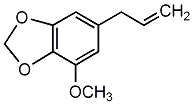
C00002762
|
Myristicin
|
CHEMBL481044
|
C005246
|
6 / 4 / 2 | 2 / 0 | No. 1917 | No. 6 |
|
Human Protein / Gene in interactions
7 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q16637 | Survival motor neuron protein | Unclassified protein | C00002762 | 4 / 1 |
| P06746 | DNA polymerase beta | Enzyme | C00002737 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00002762 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002762 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002762 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002762 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002762 | 0 / 0 |
2 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00002762
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00002762
|